Tanya Liyaqat
AgriLens: Semantic Retrieval in Agricultural Texts Using Topic Modeling and Language Models
Jan 13, 2026Abstract:As the volume of unstructured text continues to grow across domains, there is an urgent need for scalable methods that enable interpretable organization, summarization, and retrieval of information. This work presents a unified framework for interpretable topic modeling, zero-shot topic labeling, and topic-guided semantic retrieval over large agricultural text corpora. Leveraging BERTopic, we extract semantically coherent topics. Each topic is converted into a structured prompt, enabling a language model to generate meaningful topic labels and summaries in a zero-shot manner. Querying and document exploration are supported via dense embeddings and vector search, while a dedicated evaluation module assesses topical coherence and bias. This framework supports scalable and interpretable information access in specialized domains where labeled data is limited.
Stacked ensemble\-based mutagenicity prediction model using multiple modalities with graph attention network
Sep 04, 2024


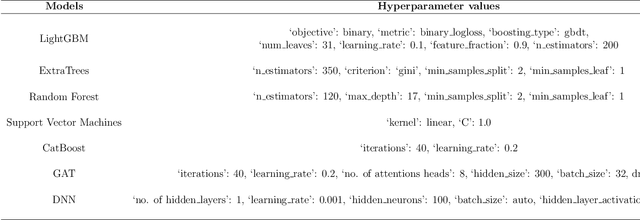
Abstract:Mutagenicity is a concern due to its association with genetic mutations which can result in a variety of negative consequences, including the development of cancer. Earlier identification of mutagenic compounds in the drug development process is therefore crucial for preventing the progression of unsafe candidates and reducing development costs. While computational techniques, especially machine learning models have become increasingly prevalent for this endpoint, they rely on a single modality. In this work, we introduce a novel stacked ensemble based mutagenicity prediction model which incorporate multiple modalities such as simplified molecular input line entry system (SMILES) and molecular graph. These modalities capture diverse information about molecules such as substructural, physicochemical, geometrical and topological. To derive substructural, geometrical and physicochemical information, we use SMILES, while topological information is extracted through a graph attention network (GAT) via molecular graph. Our model uses a stacked ensemble of machine learning classifiers to make predictions using these multiple features. We employ the explainable artificial intelligence (XAI) technique SHAP (Shapley Additive Explanations) to determine the significance of each classifier and the most relevant features in the prediction. We demonstrate that our method surpasses SOTA methods on two standard datasets across various metrics. Notably, we achieve an area under the curve of 95.21\% on the Hansen benchmark dataset, affirming the efficacy of our method in predicting mutagenicity. We believe that this research will captivate the interest of both clinicians and computational biologists engaged in translational research.
Advancements in Molecular Property Prediction: A Survey of Single and Multimodal Approaches
Aug 22, 2024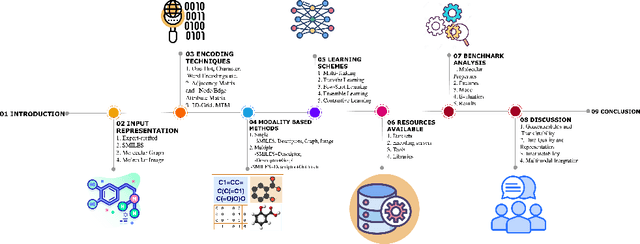
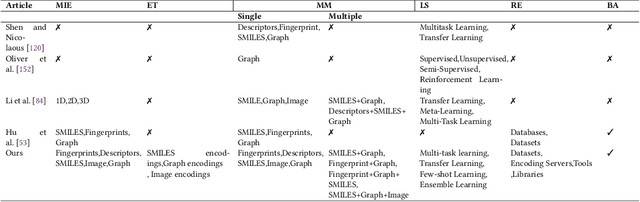
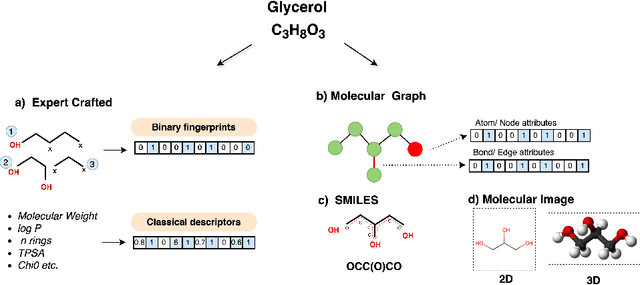

Abstract:Molecular Property Prediction (MPP) plays a pivotal role across diverse domains, spanning drug discovery, material science, and environmental chemistry. Fueled by the exponential growth of chemical data and the evolution of artificial intelligence, recent years have witnessed remarkable strides in MPP. However, the multifaceted nature of molecular data, such as molecular structures, SMILES notation, and molecular images, continues to pose a fundamental challenge in its effective representation. To address this, representation learning techniques are instrumental as they acquire informative and interpretable representations of molecular data. This article explores recent AI/-based approaches in MPP, focusing on both single and multiple modality representation techniques. It provides an overview of various molecule representations and encoding schemes, categorizes MPP methods by their use of modalities, and outlines datasets and tools available for feature generation. The article also analyzes the performance of recent methods and suggests future research directions to advance the field of MPP.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge