Tal Ashuach
Modeling variable guide efficiency in pooled CRISPR screens with ContrastiveVI+
Nov 11, 2024
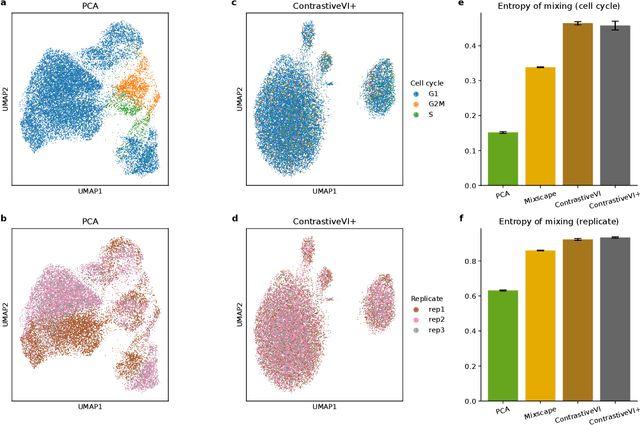
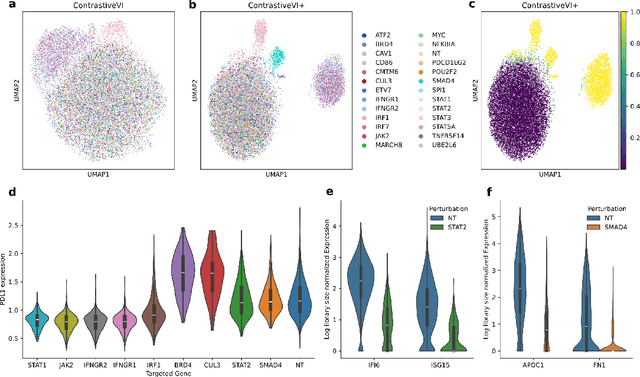
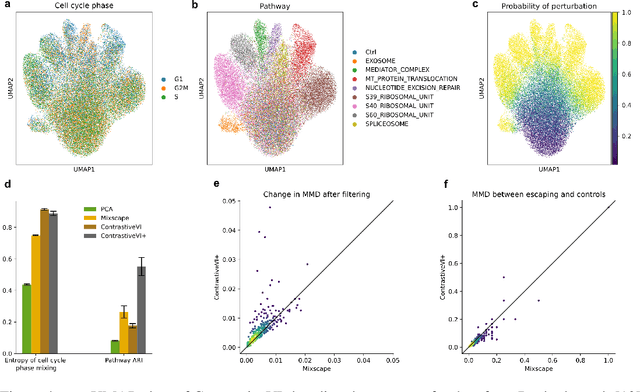
Abstract:Genetic screens mediated via CRISPR-Cas9 combined with high-content readouts have emerged as powerful tools for biological discovery. However, computational analyses of these screens come with additional challenges beyond those found with standard scRNA-seq analyses. For example, perturbation-induced variations of interest may be subtle and masked by other dominant source of variation shared with controls, and variable guide efficiency results in some cells not undergoing genetic perturbation despite expressing a guide RNA. While a number of methods have been developed to address the former problem by explicitly disentangling perturbation-induced variations from those shared with controls, less attention has been paid to the latter problem of noisy perturbation labels. To address this issue, here we propose ContrastiveVI+, a generative modeling framework that both disentangles perturbation-induced from non-perturbation-related variations while also inferring whether cells truly underwent genomic edits. Applied to three large-scale Perturb-seq datasets, we find that ContrastiveVI+ better recovers known perturbation-induced variations compared to previous methods while successfully identifying cells that escaped the functional consequences of guide RNA expression. An open-source implementation of our model is available at \url{https://github.com/insitro/contrastive_vi_plus}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge