Tajwar A. Aleef
Fast PET Scan Tumor Segmentation using Superpixels, Principal Component Analysis and K-means Clustering
Oct 18, 2017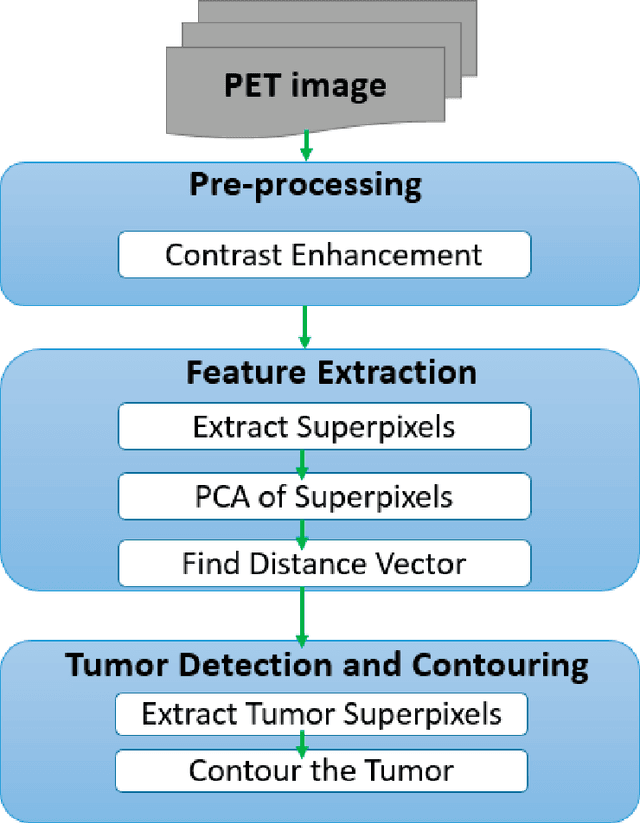


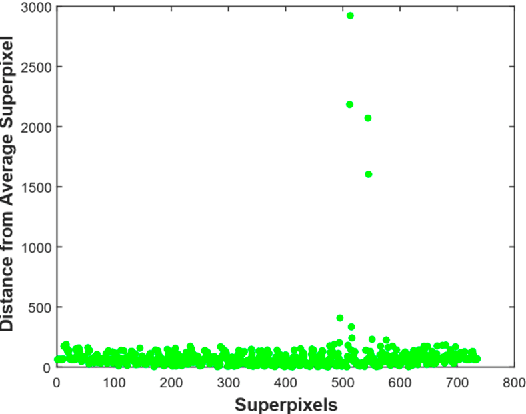
Abstract:Positron Emission Tomography scan images are extensively used in radiotherapy planning, clinical diagnosis, assessment of growth and treatment of a tumor. These all rely on fidelity and speed of detection and delineation algorithm. Despite intensive research, segmentation remained a challenging problem due to the diverse image content, resolution, shape, and noise. This paper presents a fast positron emission tomography tumor segmentation method in which superpixels are extracted first from the input image. Principal component analysis is then applied on the superpixels and also on their average. Distance vector of each superpixel from the average is computed in principal components coordinate system. Finally, k-means clustering is applied on distance vector to recognize tumor and non-tumor superpixels. The proposed approach is implemented in MATLAB 2016 which resulted in an average Dice similarity of 84.2% on the dataset. Additionally, a very fast execution time was achieved as the number of superpixels and the size of distance vector on which clustering was done was very small compared to the number of raw pixels in dataset images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge