Sumam Francis
Text Augmentations with R-drop for Classification of Tweets Self Reporting Covid-19
Nov 06, 2023Abstract:This paper presents models created for the Social Media Mining for Health 2023 shared task. Our team addressed the first task, classifying tweets that self-report Covid-19 diagnosis. Our approach involves a classification model that incorporates diverse textual augmentations and utilizes R-drop to augment data and mitigate overfitting, boosting model efficacy. Our leading model, enhanced with R-drop and augmentations like synonym substitution, reserved words, and back translations, outperforms the task mean and median scores. Our system achieves an impressive F1 score of 0.877 on the test set.
Injecting Categorical Labels and Syntactic Information into Biomedical NER
Nov 06, 2023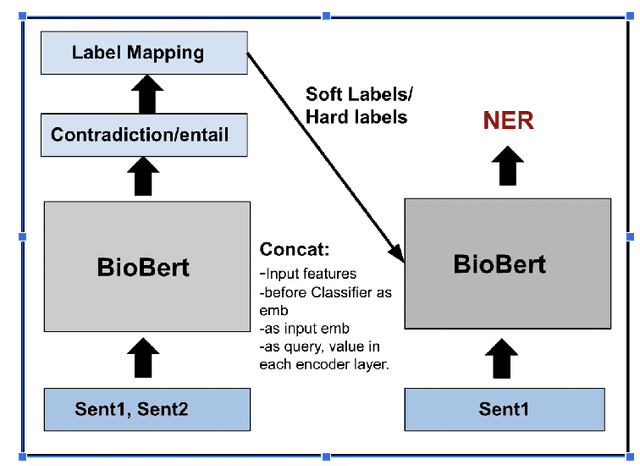
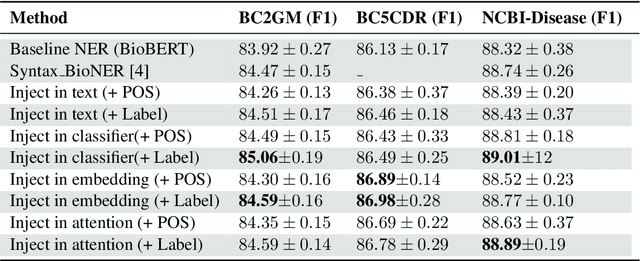
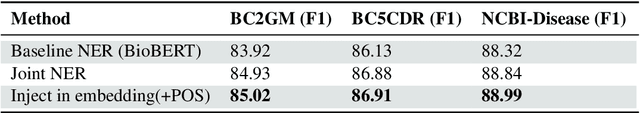
Abstract:We present a simple approach to improve biomedical named entity recognition (NER) by injecting categorical labels and Part-of-speech (POS) information into the model. We use two approaches, in the first approach, we first train a sequence-level classifier to classify the sentences into categories to obtain the sentence-level tags (categorical labels). The sequence classifier is modeled as an entailment problem by modifying the labels as a natural language template. This helps to improve the accuracy of the classifier. Further, this label information is injected into the NER model. In this paper, we demonstrate effective ways to represent and inject these labels and POS attributes into the NER model. In the second approach, we jointly learn the categorical labels and NER labels. Here we also inject the POS tags into the model to increase the syntactic context of the model. Experiments on three benchmark datasets show that incorporating categorical label information with syntactic context is quite useful and outperforms baseline BERT-based models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge