Stefan Lenz
Can open source large language models be used for tumor documentation in Germany? -- An evaluation on urological doctors' notes
Jan 21, 2025



Abstract:Tumor documentation in Germany is largely done manually, requiring reading patient records and entering data into structured databases. Large language models (LLMs) could potentially enhance this process by improving efficiency and reliability. This evaluation tests eleven different open source LLMs with sizes ranging from 1-70 billion model parameters on three basic tasks of the tumor documentation process: identifying tumor diagnoses, assigning ICD-10 codes, and extracting the date of first diagnosis. For evaluating the LLMs on these tasks, a dataset of annotated text snippets based on anonymized doctors' notes from urology was prepared. Different prompting strategies were used to investigate the effect of the number of examples in few-shot prompting and to explore the capabilities of the LLMs in general. The models Llama 3.1 8B, Mistral 7B, and Mistral NeMo 12 B performed comparably well in the tasks. Models with less extensive training data or having fewer than 7 billion parameters showed notably lower performance, while larger models did not display performance gains. Examples from a different medical domain than urology could also improve the outcome in few-shot prompting, which demonstrates the ability of LLMs to handle tasks needed for tumor documentation. Open source LLMs show a strong potential for automating tumor documentation. Models from 7-12 billion parameters could offer an optimal balance between performance and resource efficiency. With tailored fine-tuning and well-designed prompting, these models might become important tools for clinical documentation in the future. The code for the evaluation is available from https://github.com/stefan-m-lenz/UroLlmEval. We also release the dataset as a new valuable resource that addresses the shortage of authentic and easily accessible benchmarks in German-language medical NLP.
The JuliaConnectoR: a functionally oriented interface for integrating Julia in R
May 13, 2020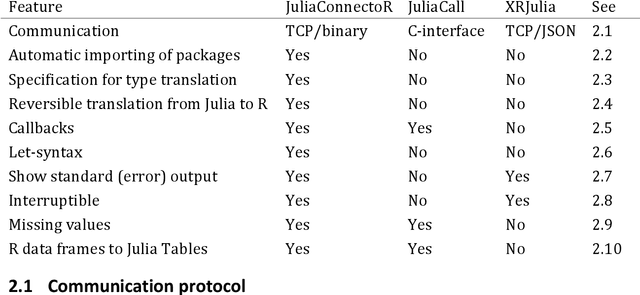
Abstract:Like many groups considering the new programming language Julia, we faced the challenge of accessing the algorithms that we develop in Julia from R. Therefore, we developed the R package JuliaConnectoR, available from the CRAN repository and GitHub (https://github.com/stefan-m-lenz/JuliaConnectoR), in particular for making advanced deep learning tools available. For maintainability and stability, we decided to base communication between R and Julia on TCP, using an optimized binary format for exchanging data. Our package also specifically contains features that allow for a convenient interactive use in R. This makes it easy to develop R extensions with Julia or to simply call functionality from Julia packages in R. With its functionally oriented design, the JuliaConnectoR enables a clean programming style by avoiding state in Julia that is not visible in the R workspace. We illustrate the further features of our package with code examples, and also discuss advantages over the two alternative packages JuliaCall and XRJulia. Finally, we demonstrate the usage of the package with a more extensive example for employing neural ordinary differential equations, a recent deep learning technique that has received much attention. This example also provides more general guidance for integrating deep learning techniques from Julia into R.
Deep generative models in DataSHIELD
Mar 11, 2020



Abstract:The best way to calculate statistics from medical data is to use the data of individual patients. In some settings, this data is difficult to obtain due to privacy restrictions. In Germany, for example, it is not possible to pool routine data from different hospitals for research purposes without the consent of the patients. The DataSHIELD software provides an infrastructure and a set of statistical methods for joint analyses of distributed data. The contained algorithms are reformulated to work with aggregated data from the participating sites instead of the individual data. If a desired algorithm is not implemented in DataSHIELD or cannot be reformulated in such a way, using artificial data is an alternative. We present a methodology together with a software implementation that builds on DataSHIELD to create artificial data that preserve complex patterns from distributed individual patient data. Such data sets of artificial patients, which are not linked to real patients, can then be used for joint analyses. We use deep Boltzmann machines (DBMs) as generative models for capturing the distribution of data. For the implementation, we employ the package "BoltzmannMachines" from the Julia programming language and wrap it for use with DataSHIELD, which is based on R. As an exemplary application, we conduct a distributed analysis with DBMs on a synthetic data set, which simulates genetic variant data. Patterns from the original data can be recovered in the artificial data using hierarchical clustering of the virtual patients, demonstrating the feasibility of the approach. Our implementation adds to DataSHIELD the ability to generate artificial data that can be used for various analyses, e. g. for pattern recognition with deep learning. This also demonstrates more generally how DataSHIELD can be flexibly extended with advanced algorithms from languages other than R.
Distributed regression modeling for selecting markers under data protection constraints
Jun 29, 2018



Abstract:Data protection constraints frequently require a distributed analysis of data, i.e., individual-level data remains at many different sites, but analysis nevertheless has to be performed jointly. The corresponding aggregated data is often exchanged manually, requiring explicit permission before transfer, i.e., the number of data calls and the amount of data should be limited. Thus, only simple aggregated summary statistics are typically transferred with just a single call. This does not allow for more complex tasks such as variable selection. As an alternative, we propose a multivariable regression approach for identifying important markers by automatic variable selection based on aggregated data from different locations in iterative calls. To minimize the amount of transferred data and the number of calls, we also provide a heuristic variant of the approach. When performing a global data standardization, the proposed methods yields the same results as when pooling individual-level data. In a simulation study, the information loss introduced by a local standardization is seen to be minimal. In a typical scenario, the heuristic decreases the number of data calls from more than 10 to 3, rendering manual data releases feasible. To make our approach widely available for application, we provide an implementation on top of the DataSHIELD framework.
Modeling Activity Tracker Data Using Deep Boltzmann Machines
Feb 28, 2018
Abstract:Commercial activity trackers are set to become an essential tool in health research, due to increasing availability in the general population. The corresponding vast amounts of mostly unlabeled data pose a challenge to statistical modeling approaches. To investigate the feasibility of deep learning approaches for unsupervised learning with such data, we examine weekly usage patterns of Fitbit activity trackers with deep Boltzmann machines (DBMs). This method is particularly suitable for modeling complex joint distributions via latent variables. We also chose this specific procedure because it is a generative approach, i.e., artificial samples can be generated to explore the learned structure. We describe how the data can be preprocessed to be compatible with binary DBMs. The results reveal two distinct usage patterns in which one group frequently uses trackers on Mondays and Tuesdays, whereas the other uses trackers during the entire week. This exemplary result shows that DBMs are feasible and can be useful for modeling activity tracker data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge