So Won Jeong
Conditional Flow Matching for Bayesian Posterior Inference
Oct 10, 2025Abstract:We propose a generative multivariate posterior sampler via flow matching. It offers a simple training objective, and does not require access to likelihood evaluation. The method learns a dynamic, block-triangular velocity field in the joint space of data and parameters, which results in a deterministic transport map from a source distribution to the desired posterior. The inverse map, named vector rank, is accessible by reversibly integrating the velocity over time. It is advantageous to leverage the dynamic design: proper constraints on the velocity yield a monotone map, which leads to a conditional Brenier map, enabling a fast and simultaneous generation of Bayesian credible sets whose contours correspond to level sets of Monge-Kantorovich data depth. Our approach is computationally lighter compared to GAN-based and diffusion-based counterparts, and is capable of capturing complex posterior structures. Finally, frequentist theoretical guarantee on the consistency of the recovered posterior distribution, and of the corresponding Bayesian credible sets, is provided.
LOBSTUR: A Local Bootstrap Framework for Tuning Unsupervised Representations in Graph Neural Networks
May 20, 2025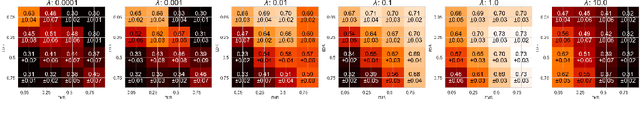
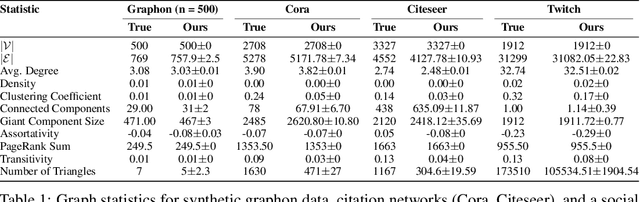
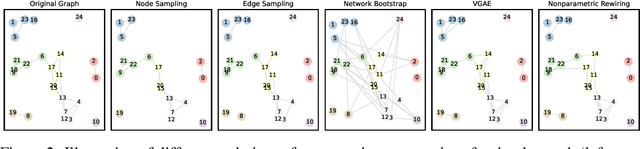
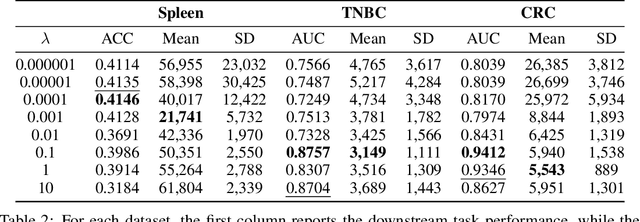
Abstract:Graph Neural Networks (GNNs) are increasingly used in conjunction with unsupervised learning techniques to learn powerful node representations, but their deployment is hindered by their high sensitivity to hyperparameter tuning and the absence of established methodologies for selecting the optimal models. To address these challenges, we propose LOBSTUR-GNN ({\bf Lo}cal {\bf B}oot{\bf s}trap for {\bf T}uning {\bf U}nsupervised {\bf R}epresentations in GNNs) i), a novel framework designed to adapt bootstrapping techniques for unsupervised graph representation learning. LOBSTUR-GNN tackles two main challenges: (a) adapting the bootstrap edge and feature resampling process to account for local graph dependencies in creating alternative versions of the same graph, and (b) establishing robust metrics for evaluating learned representations without ground-truth labels. Using locally bootstrapped resampling and leveraging Canonical Correlation Analysis (CCA) to assess embedding consistency, LOBSTUR provides a principled approach for hyperparameter tuning in unsupervised GNNs. We validate the effectiveness and efficiency of our proposed method through extensive experiments on established academic datasets, showing an 65.9\% improvement in the classification accuracy compared to an uninformed selection of hyperparameters. Finally, we deploy our framework on a real-world application, thereby demonstrating its validity and practical utility in various settings. \footnote{The code is available at \href{https://github.com/sowonjeong/lobstur-graph-bootstrap}{github.com/sowonjeong/lobstur-graph-bootstrap}.}
GNUMAP: A Parameter-Free Approach to Unsupervised Dimensionality Reduction via Graph Neural Networks
Jul 30, 2024



Abstract:With the proliferation of Graph Neural Network (GNN) methods stemming from contrastive learning, unsupervised node representation learning for graph data is rapidly gaining traction across various fields, from biology to molecular dynamics, where it is often used as a dimensionality reduction tool. However, there remains a significant gap in understanding the quality of the low-dimensional node representations these methods produce, particularly beyond well-curated academic datasets. To address this gap, we propose here the first comprehensive benchmarking of various unsupervised node embedding techniques tailored for dimensionality reduction, encompassing a range of manifold learning tasks, along with various performance metrics. We emphasize the sensitivity of current methods to hyperparameter choices -- highlighting a fundamental issue as to their applicability in real-world settings where there is no established methodology for rigorous hyperparameter selection. Addressing this issue, we introduce GNUMAP, a robust and parameter-free method for unsupervised node representation learning that merges the traditional UMAP approach with the expressivity of the GNN framework. We show that GNUMAP consistently outperforms existing state-of-the-art GNN embedding methods in a variety of contexts, including synthetic geometric datasets, citation networks, and real-world biomedical data -- making it a simple but reliable dimensionality reduction tool.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge