Shengze Dong
White-Box Diffusion Transformer for single-cell RNA-seq generation
Nov 16, 2024



Abstract:As a powerful tool for characterizing cellular subpopulations and cellular heterogeneity, single cell RNA sequencing (scRNA-seq) technology offers advantages of high throughput and multidimensional analysis. However, the process of data acquisition is often constrained by high cost and limited sample availability. To overcome these limitations, we propose a hybrid model based on Diffusion model and White-Box transformer that aims to generate synthetic and biologically plausible scRNA-seq data. Diffusion model progressively introduce noise into the data and then recover the original data through a denoising process, a forward and reverse process that is particularly suitable for generating complex data distributions. White-Box transformer is a deep learning architecture that emphasizes mathematical interpretability. By minimizing the encoding rate of the data and maximizing the sparsity of the representation, it not only reduces the computational burden, but also provides clear insight into underlying structure. Our White-Box Diffusion Transformer combines the generative capabilities of Diffusion model with the mathematical interpretability of White-Box transformer. Through experiments using six different single-cell RNA-Seq datasets, we visualize both generated and real data using t-SNE dimensionality reduction technique, as well as quantify similarity between generated and real data using various metrics to demonstrate comparable performance of White-Box Diffusion Transformer and Diffusion Transformer in generating scRNA-seq data alongside significant improvements in training efficiency and resource utilization. Our code is available at https://github.com/lingximamo/White-Box-Diffusion-Transformer
scRDiT: Generating single-cell RNA-seq data by diffusion transformers and accelerating sampling
Apr 09, 2024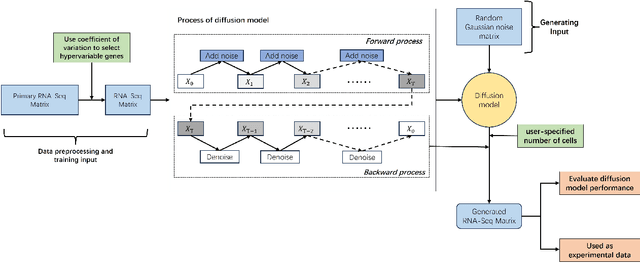
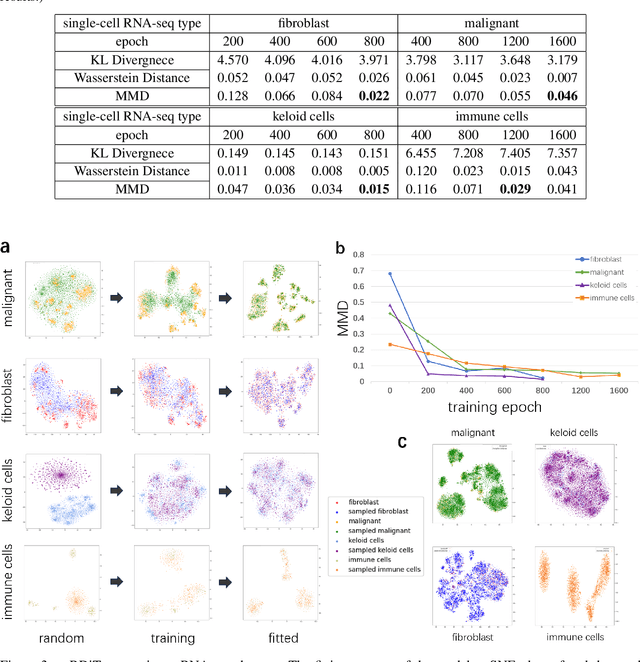
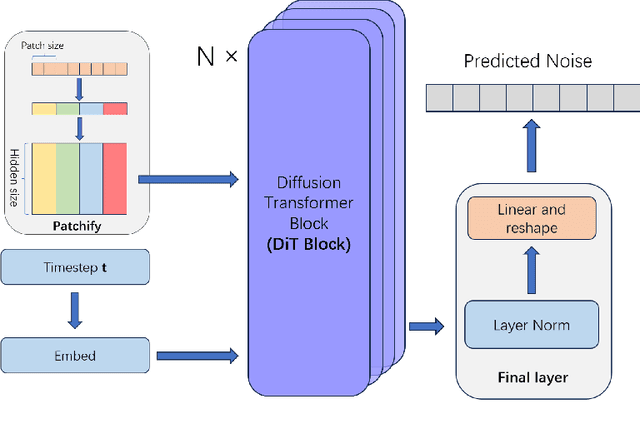
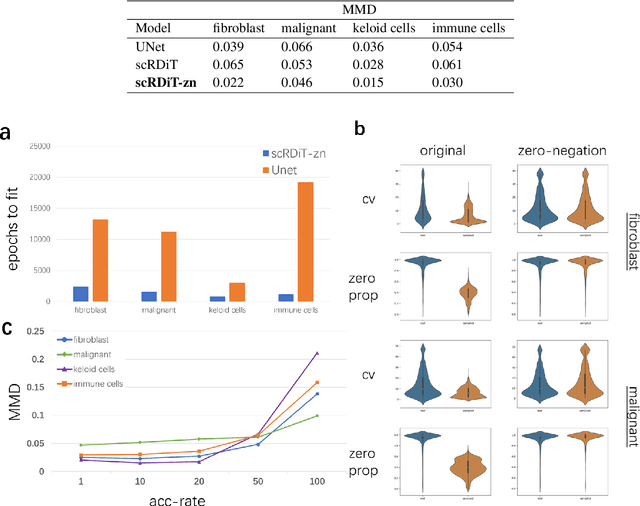
Abstract:Motivation: Single-cell RNA sequencing (scRNA-seq) is a groundbreaking technology extensively utilized in biological research, facilitating the examination of gene expression at the individual cell level within a given tissue sample. While numerous tools have been developed for scRNA-seq data analysis, the challenge persists in capturing the distinct features of such data and replicating virtual datasets that share analogous statistical properties. Results: Our study introduces a generative approach termed scRNA-seq Diffusion Transformer (scRDiT). This method generates virtual scRNA-seq data by leveraging a real dataset. The method is a neural network constructed based on Denoising Diffusion Probabilistic Models (DDPMs) and Diffusion Transformers (DiTs). This involves subjecting Gaussian noises to the real dataset through iterative noise-adding steps and ultimately restoring the noises to form scRNA-seq samples. This scheme allows us to learn data features from actual scRNA-seq samples during model training. Our experiments, conducted on two distinct scRNA-seq datasets, demonstrate superior performance. Additionally, the model sampling process is expedited by incorporating Denoising Diffusion Implicit Models (DDIM). scRDiT presents a unified methodology empowering users to train neural network models with their unique scRNA-seq datasets, enabling the generation of numerous high-quality scRNA-seq samples. Availability and implementation: https://github.com/DongShengze/scRDiT
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge