Seyed Shahim Vedaei
Multiclass Anomaly Detection in GI Endoscopic Images using Optimized Deep One-class Classification in an Imbalanced Dataset
Mar 15, 2021



Abstract:Wireless Capsule Endoscopy helps physicians examine the gastrointestinal (GI) tract noninvasively, with the cost of generating many images. Many available datasets, such as KID2 and Kvasir, suffer from imbalance issue which make it difficult to train an effective artificial intelligence (AI) system. Moreover, increasing number of classes makes the problem worse. In this study, an ensemble of one-class classifiers is used for detecting anomaly. This method focuses on learning single models using samples from only one class, and ensemble all models for multiclass classification. A total of 1,778 normal, 227 inflammation, 303 vascular diseases, and 44 polyp images have been used from the KID2 dataset. In the first step, deep features are extracted based on an autoencoder architecture from the preprocessed images. Then, these features are oversampled using Synthetic Minority Over-sampling Technique and clustered using Ordering Points to Identify the Clustering Structure. To create one-class classification model, the Support Vector Data Descriptions are trained on each cluster with the help of Ant Colony Optimization, which is also used for tuning clustering parameters for improving F1-score. This process is applied on each classes and ensemble of final models used for multiclass classification. The entire algorithm ran 5 times and obtained F1-score 96.3 +- 0.2% and macro-average F1-score 85.0 +- 0.4%, for anomaly detection and multiclass classification, respectively. The results are compared with GoogleNet, AlexNet, Resnet50, VGG16 and other published algorithms, and demonstrate that the proposed method is a competitive choice for multiclass class anomaly detection in GI images.
Siamese Network Features for Endoscopy Image and Video Localization
Mar 15, 2021



Abstract:Conventional Endoscopy (CE) and Wireless Capsule Endoscopy (WCE) are known tools for diagnosing gastrointestinal (GI) tract disorders. Localizing frames provide valuable information about the anomaly location and also can help clinicians determine a more appropriate treatment plan. There are many automated algorithms to detect the anomaly. However, very few of the existing works address the issue of localization. In this study, we present a combination of meta-learning and deep learning for localizing both endoscopy images and video. A dataset is collected from 10 different anatomical positions of human GI tract. In the meta-learning section, the system was trained using 78 CE and 27 WCE annotated frames with a modified Siamese Neural Network (SNN) to predict the location of one single image/frame. Then, a postprocessing section using bidirectional long short-term memory is proposed for localizing a sequence of frames. Here, we have employed feature vector, distance and predicted location obtained from a trained SNN. The postprocessing section is trained and tested on 1,028 and 365 seconds of CE and WCE videos using hold-out validation (50%), and achieved F1-score of 86.3% and 83.0%, respectively. In addition, we performed subjective evaluation using nine gastroenterologists. The results show that the computer-aided methods can outperform gastroenterologists assessment of localization. The proposed method is compared with various approaches, such as support vector machine with hand-crafted features, convolutional neural network and the transfer learning-based methods, and showed better results. Therefore, it can be used in frame localization, which can help in video summarization and anomaly detection.
Fetal ECG Extraction from Maternal ECG using attention-based Asymmetric CycleGAN
Nov 22, 2020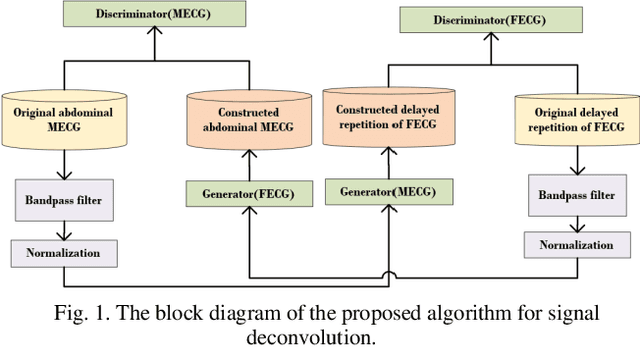
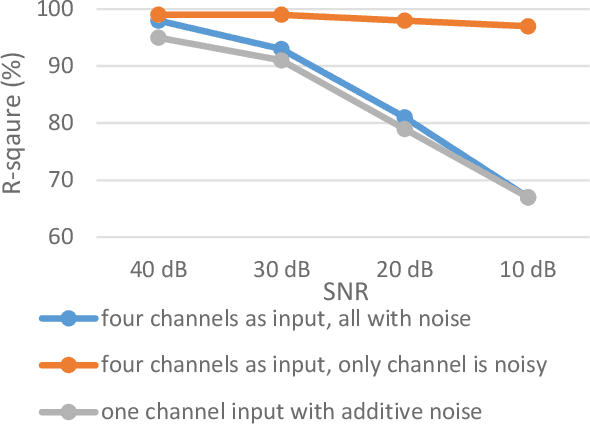
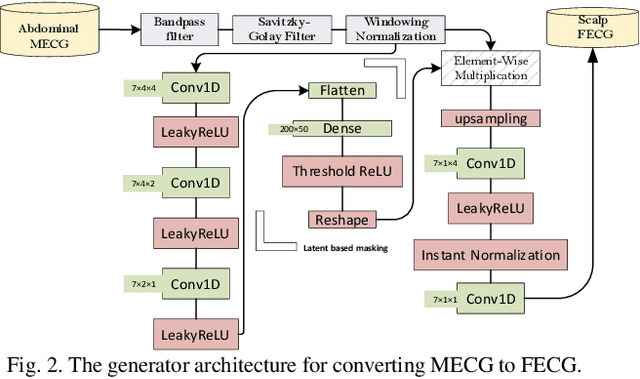
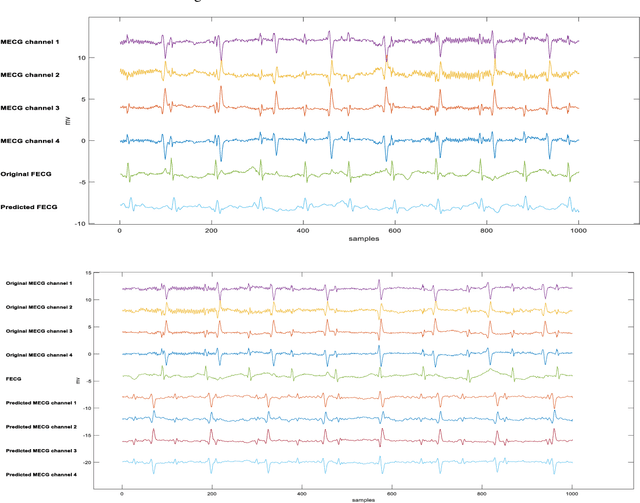
Abstract:Non-invasive fetal electrocardiogram (FECG) is used to monitor the electrical pulse of the fetal heart. Decomposing the FECG signal from maternal ECG (MECG) is a challenging problem due to the low amplitude of FECG, the overlap of R waves, and the potential exposure to noise from different sources. Traditional decomposition techniques, such as adaptive filters, require tuning, alignment, or pre-configuration, such as modeling the noise or desired signal. In this paper, a modified Cycle Generative Adversarial Network (CycleGAN) is introduced to map signal domains efficiently. The high correlation between maternal and fetal ECG parts decreases the performance of convolution layers. Therefore, masking attention layer which is inspired by the latent vector is implemented to improve generators. Three available datasets from the Physionet, including A&D FECG, NI-FECG and NI-FECG challenge, and one synthetic dataset using FECGSYN toolbox are used for evaluating the performance. The proposed method could map abdominal MECG to scalp FECG with an average 97.2% R-Square [CI 95%: 97.1, 97.2] and 7.8 +- 1.9 [CI 95% 6.13-9.47] Wavelet Energy based Diagnostic Distortion on A&D FECG dataset. Moreover, it achieved 99.4% [CI 95%: 97.8, 99.6], 99.3% [CI 95%: 97.5, 99.5] and 97.2% [CI 95%:93.3%, 97.1%] F1-score for QRS detection in A&D FECG, NI-FECG and NI-FECG challenge datasets, respectively. Finally, the generated synthetic dataset is used for investigating the effect of maternal and fetal heart rates on the proposed method. These results are comparable to the-state-of-the-art and has thus a potential of being a new algorithm for FECG extraction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge