Sebastiaan Koekkoek
Deconvolution of the Functional Ultrasound Response in the Mouse Visual Pathway Using Block-Term Decomposition
Apr 07, 2022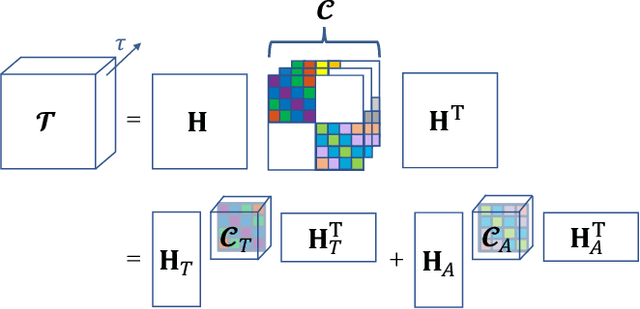
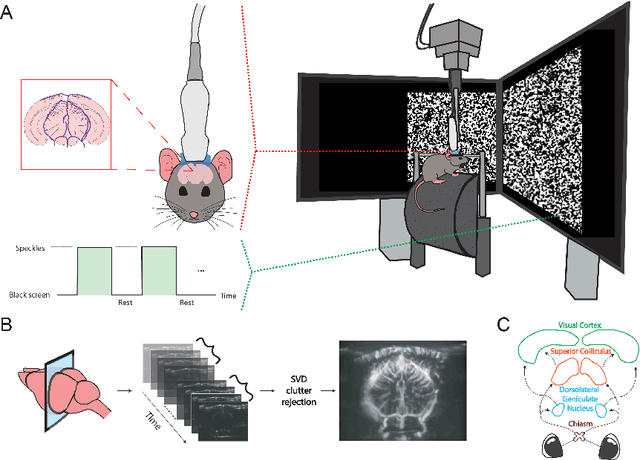
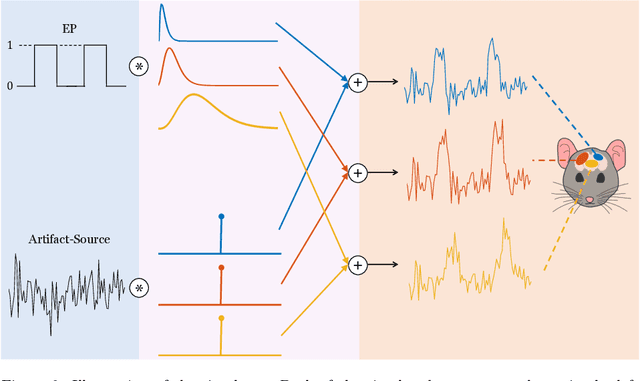
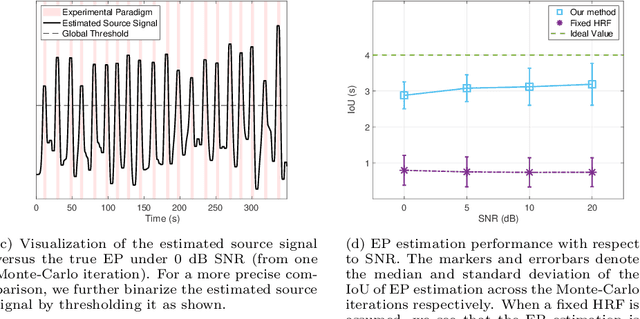
Abstract:Functional ultrasound (fUS) indirectly measures brain activity by recording changes in cerebral blood volume and flow in response to neural activation. Conventional approaches model such functional neuroimaging data as the convolution between an impulse response, known as the hemodynamic response function (HRF), and a binarized representation of the input (i.e., source) signal based on the stimulus onsets, the so-called experimental paradigm (EP). However, the EP may not be enough to characterize the whole complexity of the underlying source signals that evoke the hemodynamic changes, such as in the case of spontaneous resting state activity. Furthermore, the HRF varies across brain areas and stimuli. To achieve an adaptable framework that can capture such dynamics and unknowns of the brain function, we propose a deconvolution method for multivariate fUS time-series that reveals both the region-specific HRFs, and the source signals that induce the hemodynamic responses in the studied regions. We start by modeling the fUS time-series as convolutive mixtures and use a tensor-based approach for deconvolution based on two assumptions: (1) HRFs are parametrizable, and (2) source signals are uncorrelated. We test our approach on fUS data acquired during a visual experiment on a mouse subject, focusing on three regions within the mouse brain's colliculo-cortical, image-forming pathway: the lateral geniculate nucleus, superior colliculus and visual cortex. The estimated HRFs in each region are in agreement with prior works, whereas the estimated source signal is observed to closely follow the EP. Yet, we note a few deviations from the EP in the estimated source signal that most likely arise due to the trial-by-trial variability of the neural response across different repetitions of the stimulus observed in the selected regions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge