Satoshi Kume
Extracting Domain-specific Concepts from Large-scale Linked Open Data
Nov 22, 2021
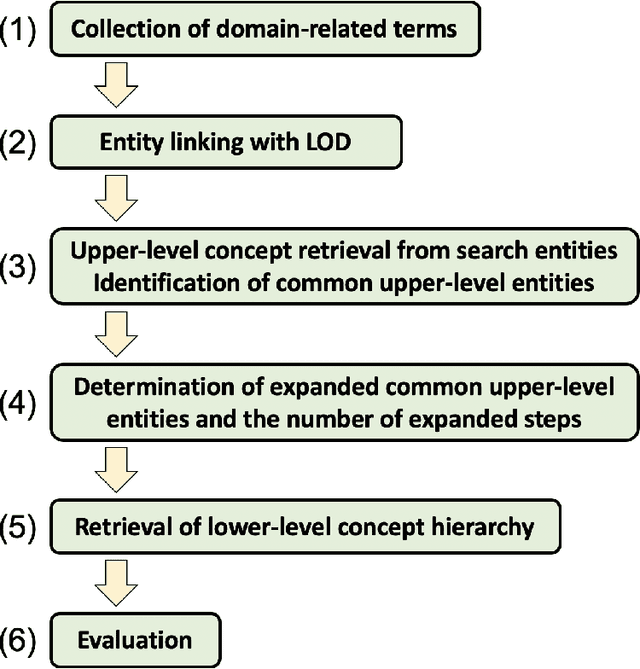
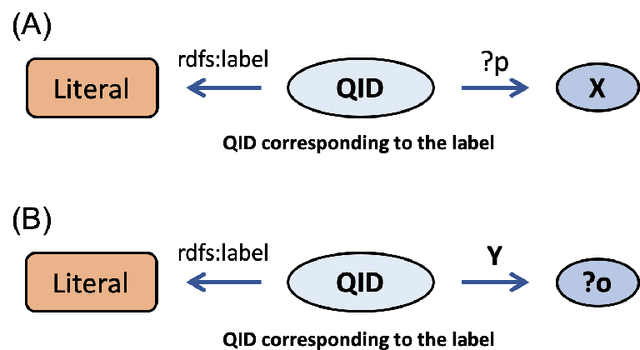
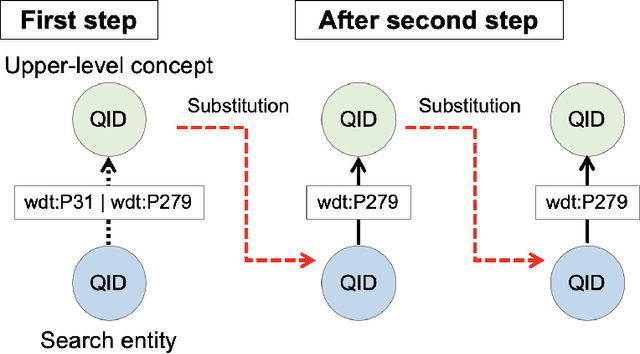
Abstract:We propose a methodology for extracting concepts for a target domain from large-scale linked open data (LOD) to support the construction of domain ontologies providing field-specific knowledge and definitions. The proposed method defines search entities by linking the LOD vocabulary with technical terms related to the target domain. The search entities are then used as a starting point for obtaining upper-level concepts in the LOD, and the occurrences of common upper-level entities and the chain-of-path relationships are examined to determine the range of conceptual connections in the target domain. A technical dictionary index and natural language processing are used to evaluate whether the extracted concepts cover the domain. As an example of extracting a class hierarchy from LOD, we used Wikidata to construct a domain ontology for polymer materials and physical properties. The proposed method can be applied to general datasets with class hierarchies, and it allows ontology developers to create an initial model of the domain ontology for their own purposes.
Development of Semantic Web-based Imaging Database for Biological Morphome
Oct 20, 2021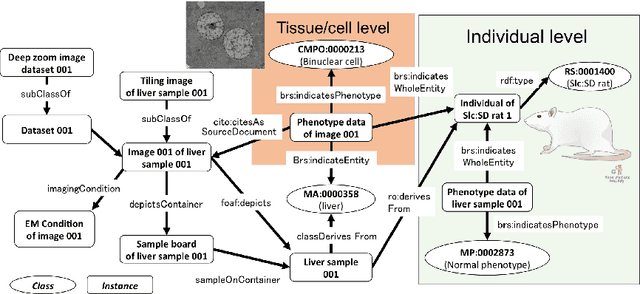
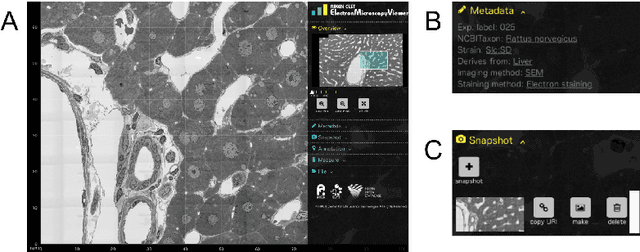
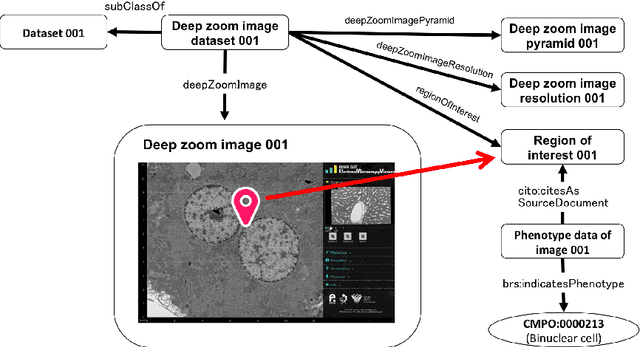
Abstract:We introduce the RIKEN Microstructural Imaging Metadatabase, a semantic web-based imaging database in which image metadata are described using the Resource Description Framework (RDF) and detailed biological properties observed in the images can be represented as Linked Open Data. The metadata are used to develop a large-scale imaging viewer that provides a straightforward graphical user interface to visualise a large microstructural tiling image at the gigabyte level. We applied the database to accumulate comprehensive microstructural imaging data produced by automated scanning electron microscopy. As a result, we have successfully managed vast numbers of images and their metadata, including the interpretation of morphological phenotypes occurring in sub-cellular components and biosamples captured in the images. We also discuss advanced utilisation of morphological imaging data that can be promoted by this database.
Development of an Ontology for an Integrated Image Analysis Platform to enable Global Sharing of Microscopy Imaging Data
Oct 20, 2021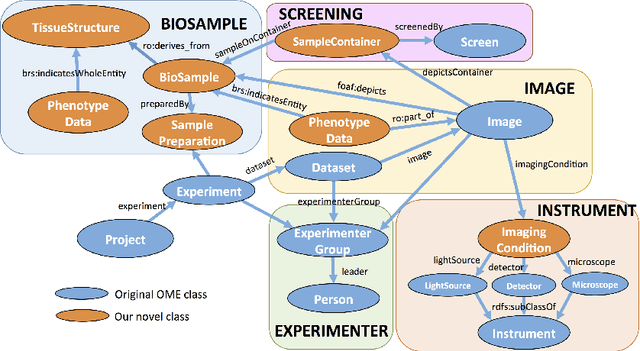
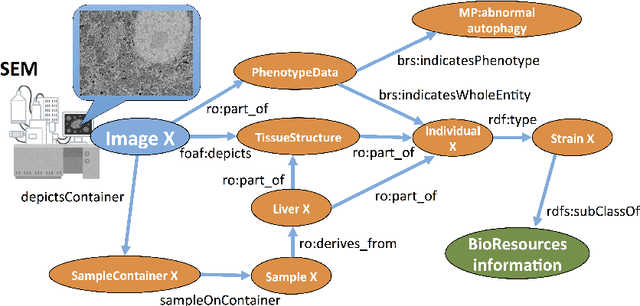
Abstract:Imaging data is one of the most important fundamentals in the current life sciences. We aimed to construct an ontology to describe imaging metadata as a data schema of the integrated database for optical and electron microscopy images combined with various bio-entities. To realise this, we applied Resource Description Framework (RDF) to an Open Microscopy Environment (OME) data model, which is the de facto standard to describe optical microscopy images and experimental data. We translated the XML-based OME metadata into the base concept of RDF schema as a trial of developing microscopy ontology. In this ontology, we propose 18 upper-level concepts including missing concepts in OME such as electron microscopy, phenotype data, biosample, and imaging conditions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge