Sarah Coffland
Emulating the Global Change Analysis Model with Deep Learning
Dec 12, 2024



Abstract:The Global Change Analysis Model (GCAM) simulates complex interactions between the coupled Earth and human systems, providing valuable insights into the co-evolution of land, water, and energy sectors under different future scenarios. Understanding the sensitivities and drivers of this multisectoral system can lead to more robust understanding of the different pathways to particular outcomes. The interactions and complexity of the coupled human-Earth systems make GCAM simulations costly to run at scale - a requirement for large ensemble experiments which explore uncertainty in model parameters and outputs. A differentiable emulator with similar predictive power, but greater efficiency, could provide novel scenario discovery and analysis of GCAM and its outputs, requiring fewer runs of GCAM. As a first use case, we train a neural network on an existing large ensemble that explores a range of GCAM inputs related to different relative contributions of energy production sources, with a focus on wind and solar. We complement this existing ensemble with interpolated input values and a wider selection of outputs, predicting 22,528 GCAM outputs across time, sectors, and regions. We report a median $R^2$ score of 0.998 for the emulator's predictions and an $R^2$ score of 0.812 for its input-output sensitivity.
RoseNet: Predicting Energy Metrics of Double InDel Mutants Using Deep Learning
Oct 20, 2023
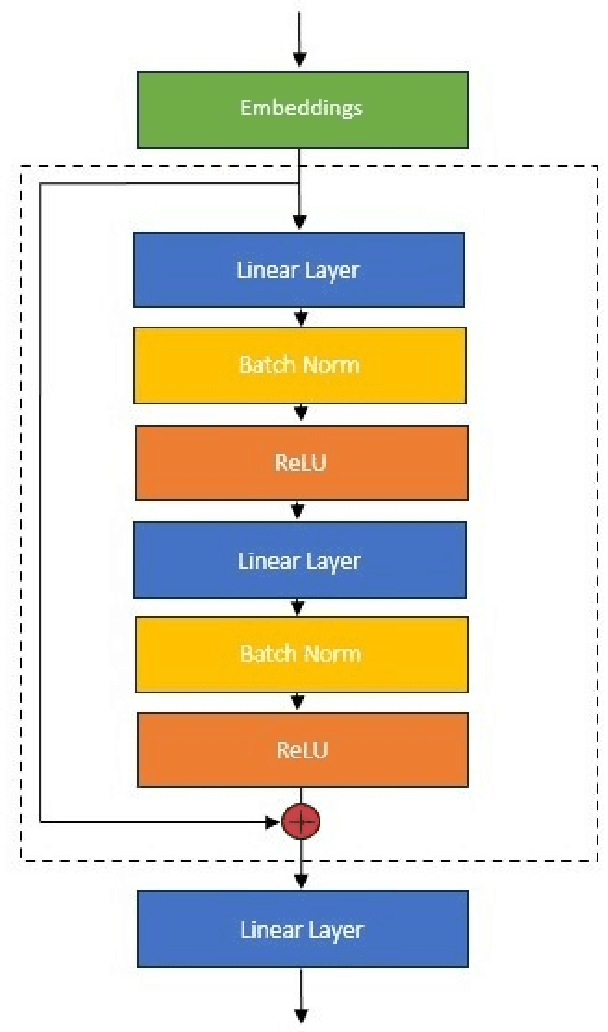
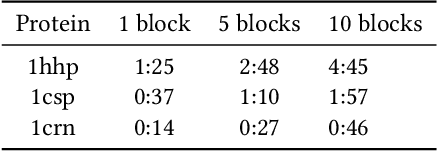
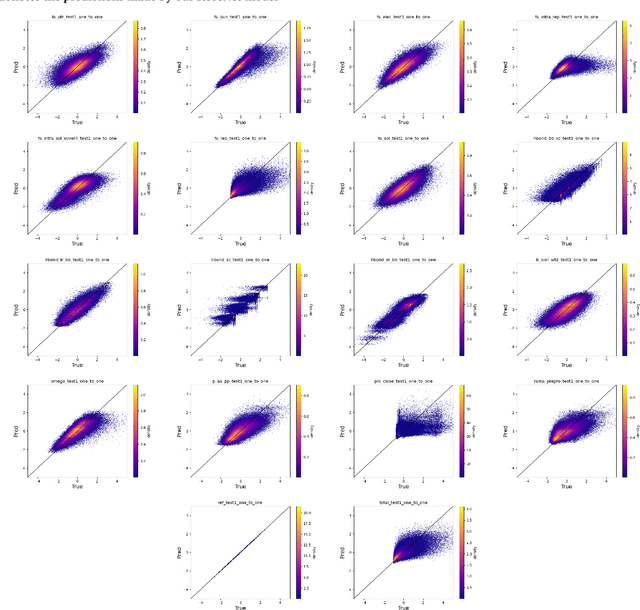
Abstract:An amino acid insertion or deletion, or InDel, can have profound and varying functional impacts on a protein's structure. InDel mutations in the transmembrane conductor regulator protein for example give rise to cystic fibrosis. Unfortunately performing InDel mutations on physical proteins and studying their effects is a time prohibitive process. Consequently, modeling InDels computationally can supplement and inform wet lab experiments. In this work, we make use of our data sets of exhaustive double InDel mutations for three proteins which we computationally generated using a robotics inspired inverse kinematics approach available in Rosetta. We develop and train a neural network, RoseNet, on several structural and energetic metrics output by Rosetta during the mutant generation process. We explore and present how RoseNet is able to emulate the exhaustive data set using deep learning methods, and show to what extent it can predict Rosetta metrics for unseen mutant sequences with two InDels. RoseNet achieves a Pearson correlation coefficient median accuracy of 0.775 over all Rosetta scores for the largest protein. Furthermore, a sensitivity analysis is performed to determine the necessary quantity of data required to accurately emulate the structural scores for computationally generated mutants. We show that the model can be trained on minimal data (<50%) and still retain a high level of accuracy.
* Presented at Computational Structural Bioinformatics Workshop 2023
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge