Sarah C. Brüningk
Interpretability Aware Model Training to Improve Robustness against Out-of-Distribution Magnetic Resonance Images in Alzheimer's Disease Classification
Nov 15, 2021
Abstract:Owing to its pristine soft-tissue contrast and high resolution, structural magnetic resonance imaging (MRI) is widely applied in neurology, making it a valuable data source for image-based machine learning (ML) and deep learning applications. The physical nature of MRI acquisition and reconstruction, however, causes variations in image intensity, resolution, and signal-to-noise ratio. Since ML models are sensitive to such variations, performance on out-of-distribution data, which is inherent to the setting of a deployed healthcare ML application, typically drops below acceptable levels. We propose an interpretability aware adversarial training regime to improve robustness against out-of-distribution samples originating from different MRI hardware. The approach is applied to 1.5T and 3T MRIs obtained from the Alzheimer's Disease Neuroimaging Initiative database. We present preliminary results showing promising performance on out-of-distribution samples.
Image analysis for Alzheimer's disease prediction: Embracing pathological hallmarks for model architecture design
Nov 13, 2020
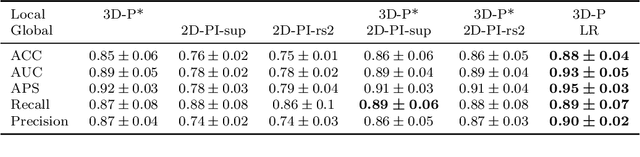
Abstract:Alzheimer's disease (AD) is associated with local (e.g. brain tissue atrophy) and global brain changes (loss of cerebral connectivity), which can be detected by high-resolution structural magnetic resonance imaging. Conventionally, these changes and their relation to AD are investigated independently. Here, we introduce a novel, highly-scalable approach that simultaneously captures $\textit{local}$ and $\textit{global}$ changes in the diseased brain. It is based on a neural network architecture that combines patch-based, high-resolution 3D-CNNs with global topological features, evaluating multi-scale brain tissue connectivity. Our local-global approach reached competitive results with an average precision score of $0.95\pm0.03$ for the classification of cognitively normal subjects and AD patients (prevalence $\approx 55\%$).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge