Rudolf Stollberger
Pfungst and Clever Hans: Identifying the unintended cues in a widely used Alzheimer's disease MRI dataset using explainable deep learning
Jan 27, 2025



Abstract:Backgrounds. Deep neural networks have demonstrated high accuracy in classifying Alzheimer's disease (AD). This study aims to enlighten the underlying black-box nature and reveal individual contributions of T1-weighted (T1w) gray-white matter texture, volumetric information and preprocessing on classification performance. Methods. We utilized T1w MRI data from the Alzheimer's Disease Neuroimaging Initiative to distinguish matched AD patients (990 MRIs) from healthy controls (990 MRIs). Preprocessing included skull stripping and binarization at varying thresholds to systematically eliminate texture information. A deep neural network was trained on these configurations, and the model performance was compared using McNemar tests with discrete Bonferroni-Holm correction. Layer-wise Relevance Propagation (LRP) and structural similarity metrics between heatmaps were applied to analyze learned features. Results. Classification performance metrics (accuracy, sensitivity, and specificity) were comparable across all configurations, indicating a negligible influence of T1w gray- and white signal texture. Models trained on binarized images demonstrated similar feature performance and relevance distributions, with volumetric features such as atrophy and skull-stripping features emerging as primary contributors. Conclusions. We revealed a previously undiscovered Clever Hans effect in a widely used AD MRI dataset. Deep neural networks classification predominantly rely on volumetric features, while eliminating gray-white matter T1w texture did not decrease the performance. This study clearly demonstrates an overestimation of the importance of gray-white matter contrasts, at least for widely used structural T1w images, and highlights potential misinterpretation of performance metrics.
Joint multi-field T$_1$ quantification for fast field-cycling MRI
Feb 19, 2021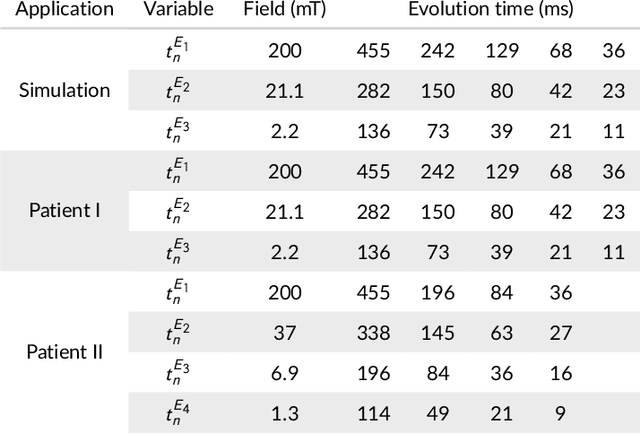
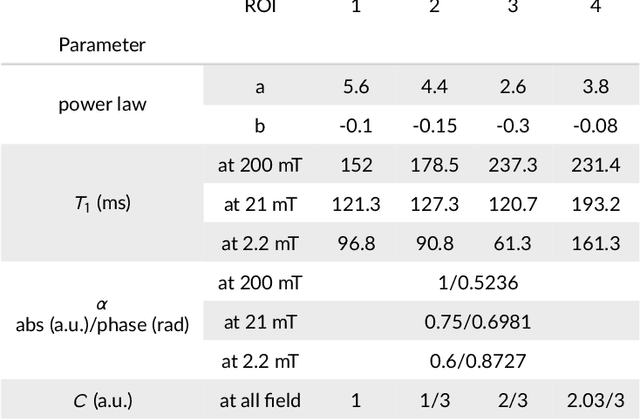
Abstract:Recent developments in hardware design enable the use of Fast Field-Cycling (FFC) techniques in MRI to exploit the different relaxation rates at very low field strength, achieving novel contrast. The method opens new avenues for in vivo characterisations of pathologies but at the expense of longer acquisition times. To mitigate this we propose a model-based reconstruction method that fully exploits the high information redundancy offered by FFC methods. This is based on joining spatial information from all fields based on TGV regularization. The algorithm was tested on brain stroke images, both simulated and acquired from FFC patients scans using an FFC spin echo sequences. The results are compared to non-linear least squares combined with k-space filtering. The proposed method shows excellent abilities to remove noise while maintaining sharp image features with large SNR gains at low-field images, clearly outperforming the reference approach. Especially patient data shows huge improvements in visual appearance over all fields. The proposed reconstruction technique largely improves FFC image quality, further pushing this new technology towards clinical standards.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge