Rohan Gala
Tuning Music Education: AI-Powered Personalization in Learning Music
Dec 18, 2024

Abstract:Recent AI-driven step-function advances in several longstanding problems in music technology are opening up new avenues to create the next generation of music education tools. Creating personalized, engaging, and effective learning experiences are continuously evolving challenges in music education. Here we present two case studies using such advances in music technology to address these challenges. In our first case study we showcase an application that uses Automatic Chord Recognition to generate personalized exercises from audio tracks, connecting traditional ear training with real-world musical contexts. In the second case study we prototype adaptive piano method books that use Automatic Music Transcription to generate exercises at different skill levels while retaining a close connection to musical interests. These applications demonstrate how recent AI developments can democratize access to high-quality music education and promote rich interaction with music in the age of generative AI. We hope this work inspires other efforts in the community, aimed at removing barriers to access to high-quality music education and fostering human participation in musical expression.
Joint Learning of Discrete and Continuous Variability with Coupled Autoencoding Agents
Jul 20, 2020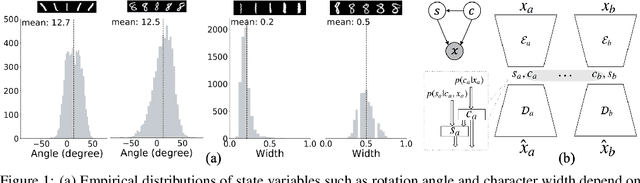
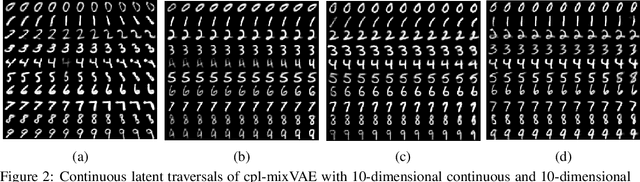
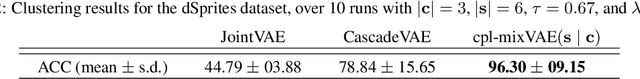
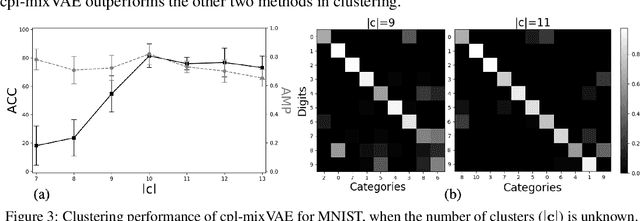
Abstract:Jointly identifying discrete and continuous factors of variability can help unravel complex phenomena. In neuroscience, a high-priority instance of this problem is the analysis of neuronal identity. Here, we study this problem in a variational framework by utilizing interacting autoencoding agents, designed to function in the absence of prior distribution over the discrete variable and reach collective decisions. We provide theoretical justification for our method and demonstrate improvements in terms of interpretability, stability, and accuracy over comparable approaches with experimental results on two benchmark datasets and a recent dataset of gene expression profiles of mouse cortical neurons. Furthermore, we demonstrate how our method can determine the neuronal cell types in an unsupervised setting, while identifying the genes implicated in regulating biologically relevant neuronal states.
A coupled autoencoder approach for multi-modal analysis of cell types
Nov 06, 2019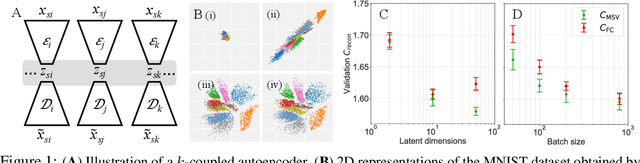
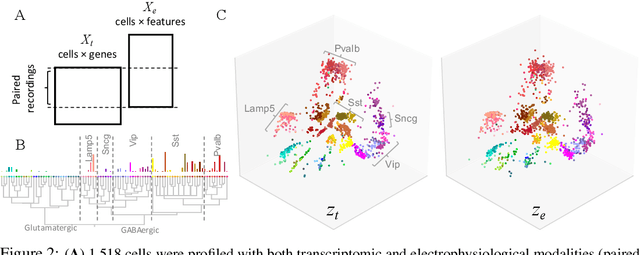
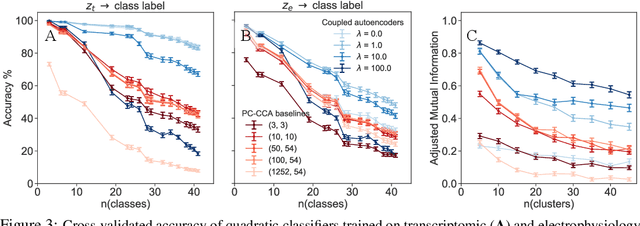
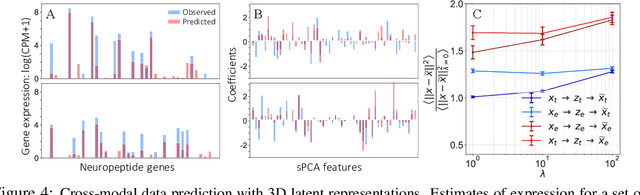
Abstract:Recent developments in high throughput profiling of individual neurons have spurred data driven exploration of the idea that there exist natural groupings of neurons referred to as cell types. The promise of this idea is that the immense complexity of brain circuits can be reduced, and effectively studied by means of interactions between cell types. While clustering of neuron populations based on a particular data modality can be used to define cell types, such definitions are often inconsistent across different characterization modalities. We pose this issue of cross-modal alignment as an optimization problem and develop an approach based on coupled training of autoencoders as a framework for such analyses. We apply this framework to a Patch-seq dataset consisting of transcriptomic and electrophysiological profiles for the same set of neurons to study consistency of representations across modalities, and evaluate cross-modal data prediction ability. We explore the problem where only a subset of neurons is characterized with more than one modality, and demonstrate that representations learned by coupled autoencoders can be used to identify types sampled only by a single modality.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge