Robert Moskovitch
The Semantic Adjacency Criterion in Time Intervals Mining
Jan 11, 2021
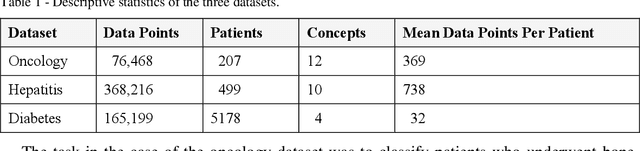
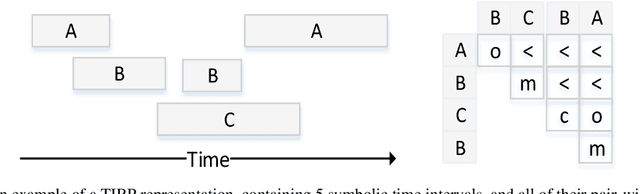
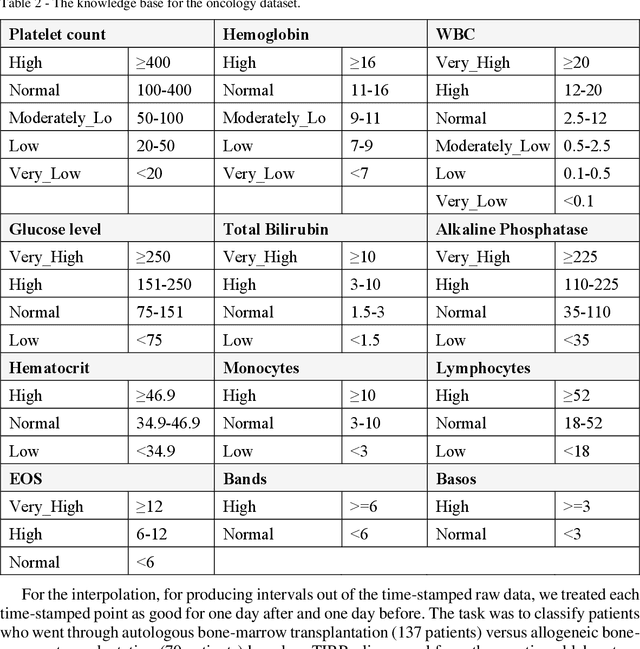
Abstract:Frequent temporal patterns discovered in time-interval-based multivariate data, although syntactically correct, might be non-transparent: For some pattern instances, there might exist intervals for the same entity that contradict the pattern's usual meaning. We conjecture that non-transparent patterns are also less useful as classification or prediction features. We propose a new pruning constraint during a frequent temporal-pattern discovery process, the Semantic Adjacency Criterion [SAC], which exploits domain knowledge to filter out patterns that contain potentially semantically contradictory components. We have defined three SAC versions, and tested their effect in three medical domains. We embedded these criteria in a frequent-temporal-pattern discovery framework. Previously, we had informally presented the SAC principle and showed that using it to prune patterns enhances the repeatability of their discovery in the same clinical domain. Here, we define formally the semantics of three SAC variations, and compare the use of the set of pruned patterns to the use of the complete set of discovered patterns, as features for classification and prediction tasks in three different medical domains. We induced four classifiers for each task, using four machine-learning methods: Random Forests, Naive Bayes, SVM, and Logistic Regression. The features were frequent temporal patterns discovered in each data set. SAC-based temporal pattern-discovery reduced by up to 97% the number of discovered patterns and by up to 98% the discovery runtime. But the classification and prediction performance of the reduced SAC-based pattern-based features set, was as good as when using the complete set. Using SAC can significantly reduce the number of discovered frequent interval-based temporal patterns, and the corresponding computational effort, without losing classification or prediction performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge