Rikin Hargunani
Interpretability and Individuality in Knee MRI: Patient-Specific Radiomic Fingerprint with Reconstructed Healthy Personas
Jan 13, 2026Abstract:For automated assessment of knee MRI scans, both accuracy and interpretability are essential for clinical use and adoption. Traditional radiomics rely on predefined features chosen at the population level; while more interpretable, they are often too restrictive to capture patient-specific variability and can underperform end-to-end deep learning (DL). To address this, we propose two complementary strategies that bring individuality and interpretability: radiomic fingerprints and healthy personas. First, a radiomic fingerprint is a dynamically constructed, patient-specific feature set derived from MRI. Instead of applying a uniform population-level signature, our model predicts feature relevance from a pool of candidate features and selects only those most predictive for each patient, while maintaining feature-level interpretability. This fingerprint can be viewed as a latent-variable model of feature usage, where an image-conditioned predictor estimates usage probabilities and a transparent logistic regression with global coefficients performs classification. Second, a healthy persona synthesises a pathology-free baseline for each patient using a diffusion model trained to reconstruct healthy knee MRIs. Comparing features extracted from pathological images against their personas highlights deviations from normal anatomy, enabling intuitive, case-specific explanations of disease manifestations. We systematically compare fingerprints, personas, and their combination across three clinical tasks. Experimental results show that both approaches yield performance comparable to or surpassing state-of-the-art DL models, while supporting interpretability at multiple levels. Case studies further illustrate how these perspectives facilitate human-explainable biomarker discovery and pathology localisation.
Patient-specific radiomic feature selection with reconstructed healthy persona of knee MR images
Mar 17, 2025


Abstract:Classical radiomic features have been designed to describe image appearance and intensity patterns. These features are directly interpretable and readily understood by radiologists. Compared with end-to-end deep learning (DL) models, lower dimensional parametric models that use such radiomic features offer enhanced interpretability but lower comparative performance in clinical tasks. In this study, we propose an approach where a standard logistic regression model performance is substantially improved by learning to select radiomic features for individual patients, from a pool of candidate features. This approach has potentials to maintain the interpretability of such approaches while offering comparable performance to DL. We also propose to expand the feature pool by generating a patient-specific healthy persona via mask-inpainting using a denoising diffusion model trained on healthy subjects. Such a pathology-free baseline feature set allows further opportunity in novel feature discovery and improved condition classification. We demonstrate our method on multiple clinical tasks of classifying general abnormalities, anterior cruciate ligament tears, and meniscus tears. Experimental results demonstrate that our approach achieved comparable or even superior performance than state-of-the-art DL approaches while offering added interpretability by using radiomic features extracted from images and supplemented by generating healthy personas. Example clinical cases are discussed in-depth to demonstrate the intepretability-enabled utilities such as human-explainable feature discovery and patient-specific location/view selection. These findings highlight the potentials of the combination of subject-specific feature selection with generative models in augmenting radiomic analysis for more interpretable decision-making. The codes are available at: https://github.com/YaxiiC/RadiomicsPersona.git
Segmentation by registration-enabled SAM prompt engineering using five reference images
Jul 25, 2024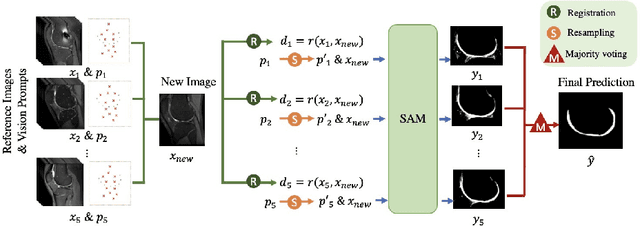
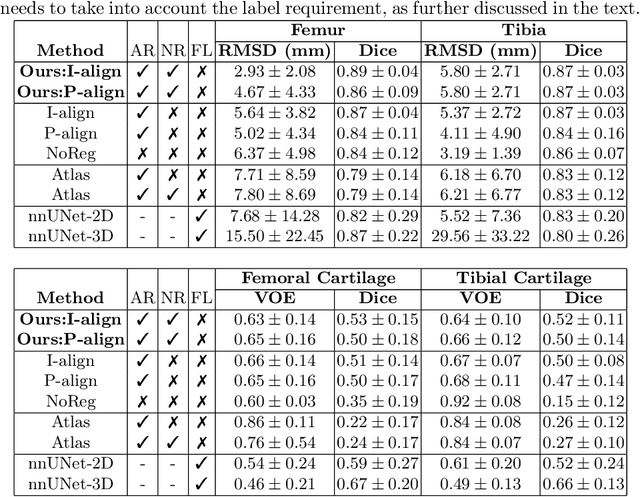
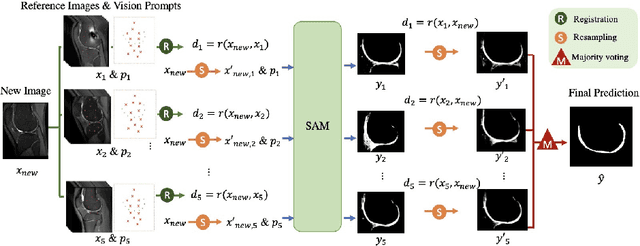
Abstract:The recently proposed Segment Anything Model (SAM) is a general tool for image segmentation, but it requires additional adaptation and careful fine-tuning for medical image segmentation, especially for small, irregularly-shaped, and boundary-ambiguous anatomical structures such as the knee cartilage that is of interest in this work. Repaired cartilage, after certain surgical procedures, exhibits imaging patterns unseen to pre-training, posing further challenges for using models like SAM with or without general-purpose fine-tuning. To address this, we propose a novel registration-based prompt engineering framework for medical image segmentation using SAM. This approach utilises established image registration algorithms to align the new image (to-be-segmented) and a small number of reference images, without requiring segmentation labels. The spatial transformations generated by registration align either the new image or pre-defined point-based prompts, before using them as input to SAM. This strategy, requiring as few as five reference images with defined point prompts, effectively prompts SAM for inference on new images, without needing any segmentation labels. Evaluation of MR images from patients who received cartilage stem cell therapy yielded Dice scores of 0.89, 0.87, 0.53, and 0.52 for segmenting femur, tibia, femoral- and tibial cartilages, respectively. This outperforms atlas-based label fusion and is comparable to supervised nnUNet, an upper-bound fair baseline in this application, both of which require full segmentation labels for reference samples. The codes are available at: https://github.com/chrissyinreallife/KneeSegmentWithSAM.git
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge