Rifat Zahan
DNA Methylation Data to Predict Suicidal and Non-Suicidal Deaths: A Machine Learning Approach
Apr 04, 2020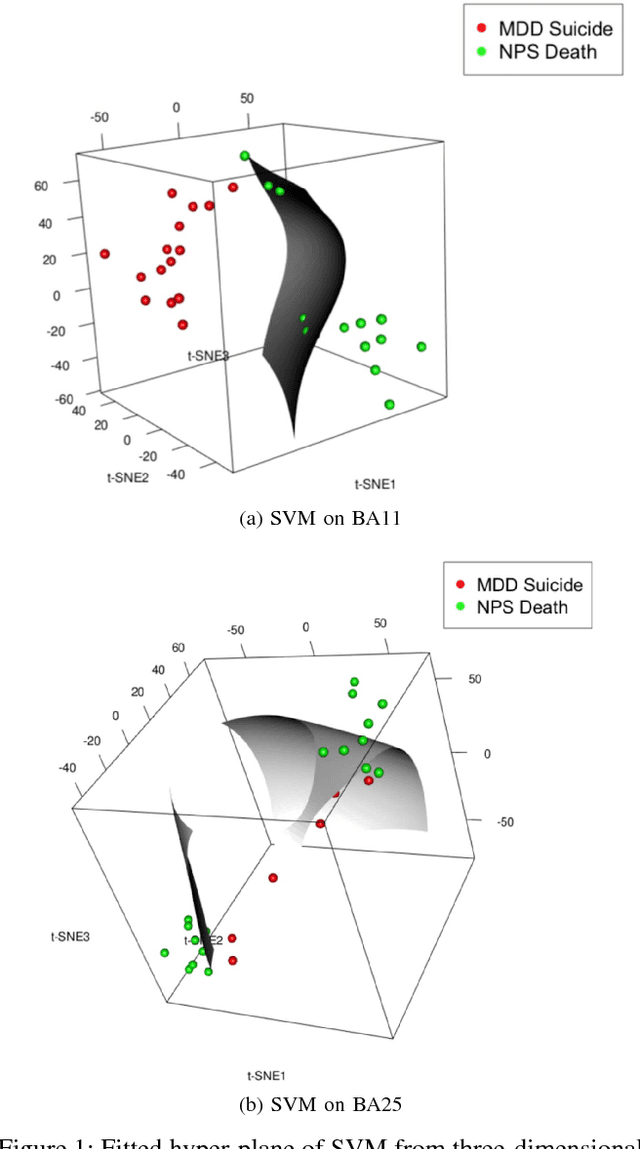
Abstract:The objective of this study is to predict suicidal and non-suicidal deaths from DNA methylation data using a modern machine learning algorithm. We used support vector machines to classify existing secondary data consisting of normalized values of methylated DNA probe intensities from tissues of two cortical brain regions to distinguish suicide cases from control cases. Before classification, we employed Principal component analysis (PCA) and t-distributed Stochastic Neighbor Embedding (t-SNE) to reduce the dimension of the data. In comparison to PCA, the modern data visualization method t-SNE performs better in dimensionality reduction. t-SNE accounts for the possible non-linear patterns in low-dimensional data. We applied four-fold cross-validation in which the resulting output from t-SNE was used as training data for the Support Vector Machine (SVM). Despite the use of cross-validation, the nominally perfect prediction of suicidal deaths for BA11 data suggests possible over-fitting of the model. The study also may have suffered from 'spectrum bias' since the individuals were only studied from two extreme scenarios. This research constitutes a baseline study for classifying suicidal and non-suicidal deaths from DNA methylation data. Future studies with larger sample size, while possibly incorporating methylation data from living individuals, may reduce the bias and improve the accuracy of the results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge