Rick Hennig
Predicting 3D RNA Folding Patterns via Quadratic Binary Optimization
Jun 14, 2021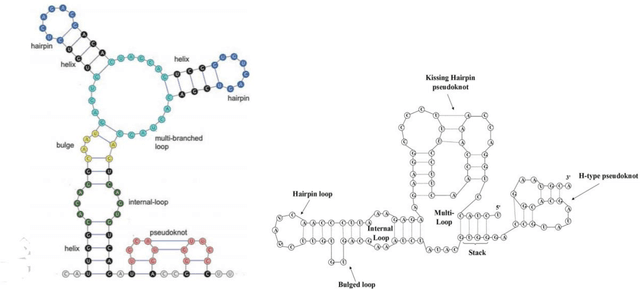
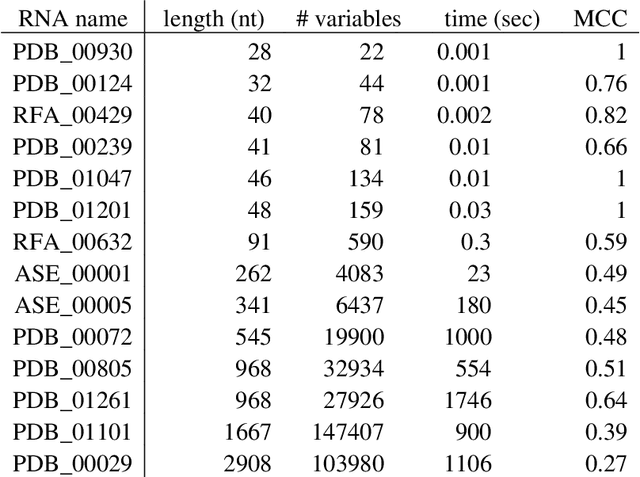
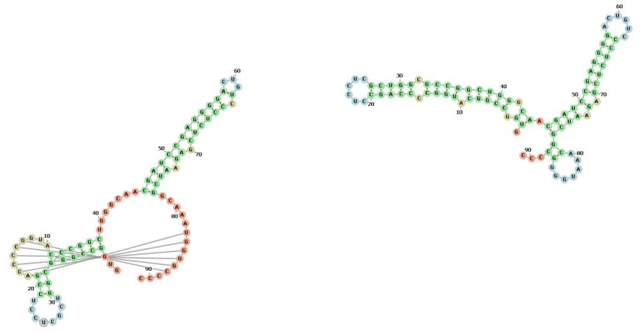

Abstract:The structure of an RNA molecule plays a significant role in its biological function. Predicting structure given a one dimensional sequence of RNA nucleotide bases is a difficult and important problem. Many computer programs (known as in silico) are available for predicting 2-dimensional (secondary) structures however 3-dimensional (tertiary) structure prediction is much more difficult mainly due to the far greater number of feasible solutions and fewer experimental data on the thermodynamic energies of 3D structures. It is also challenging to verify the most likely three dimensional structure even with the availability of sophisticated x-ray crystallography and nuclear magnetic resonance imaging technologies. In this paper we develop three dimensional RNA folding predictions by adding penalty and reward parameters to a previous two dimensional approach based on Quadratic Unconstrained Binary Optimization (QUBO) models. These parameters provide flexibility in the amount of three dimensional folding allowed. We address the problem of multiple near-optimal structures via a new weighted similarity structure measure and illustrate folding pathways via progressively improving local optimal solutions. The problems are solved via a new commercial QUBO solver AlphaQUBO (Meta-Analytics, 2020) that solves problems having hundreds of thousands of binary variables.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge