Richard Threlfall
Molecular Generation with Recurrent Neural Networks (RNNs)
May 17, 2017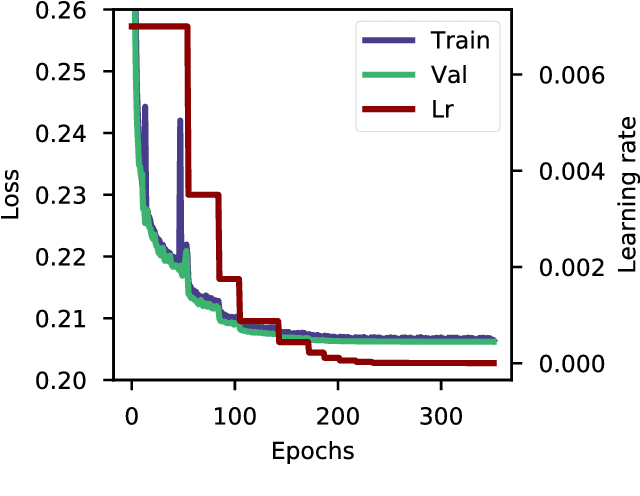
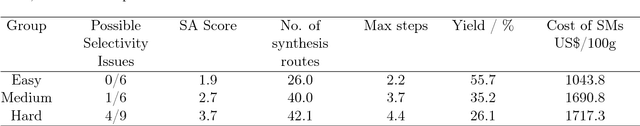
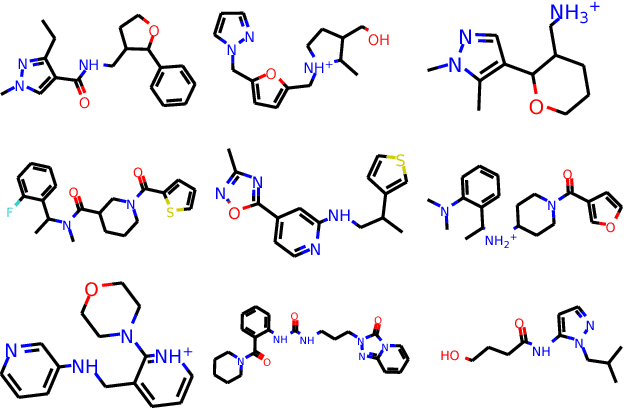
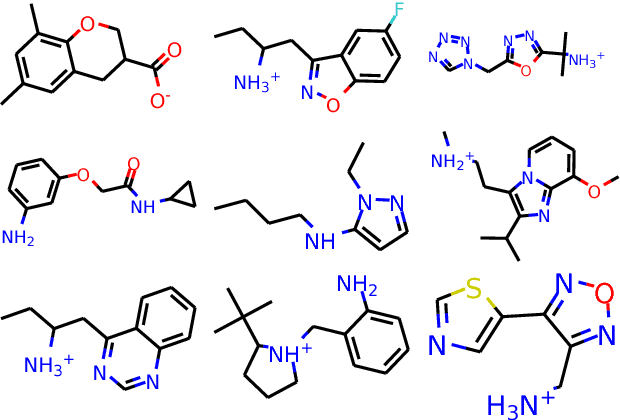
Abstract:The potential number of drug like small molecules is estimated to be between 10^23 and 10^60 while current databases of known compounds are orders of magnitude smaller with approximately 10^8 compounds. This discrepancy has led to an interest in generating virtual libraries using hand crafted chemical rules and fragment based methods to cover a larger area of chemical space and generate chemical libraries for use in in silico drug discovery endeavors. Here it is explored to what extent a recurrent neural network with long short term memory cells can figure out sensible chemical rules and generate synthesizable molecules by being trained on existing compounds encoded as SMILES. The networks can to a high extent generate novel, but chemically sensible molecules. The properties of the molecules are tuned by training on two different datasets consisting of fragment like molecules and drug like molecules. The produced molecules and the training databases have very similar distributions of molar weight, predicted logP, number of hydrogen bond acceptors and donors, number of rotatable bonds and topological polar surface area when compared to their respective training sets. The compounds are for the most cases synthesizable as assessed with SA score and Wiley ChemPlanner.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge