Rafał Dreżewski
PSO-UNet: Particle Swarm-Optimized U-Net Framework for Precise Multimodal Brain Tumor Segmentation
Mar 24, 2025Abstract:Medical image segmentation, particularly for brain tumor analysis, demands precise and computationally efficient models due to the complexity of multimodal MRI datasets and diverse tumor morphologies. This study introduces PSO-UNet, which integrates Particle Swarm Optimization (PSO) with the U-Net architecture for dynamic hyperparameter optimization. Unlike traditional manual tuning or alternative optimization approaches, PSO effectively navigates complex hyperparameter search spaces, explicitly optimizing the number of filters, kernel size, and learning rate. PSO-UNet substantially enhances segmentation performance, achieving Dice Similarity Coefficients (DSC) of 0.9578 and 0.9523 and Intersection over Union (IoU) scores of 0.9194 and 0.9097 on the BraTS 2021 and Figshare datasets, respectively. Moreover, the method reduces computational complexity significantly, utilizing only 7.8 million parameters and executing in approximately 906 seconds, markedly faster than comparable U-Net-based frameworks. These outcomes underscore PSO-UNet's robust generalization capabilities across diverse MRI modalities and tumor classifications, emphasizing its clinical potential and clear advantages over conventional hyperparameter tuning methods. Future research will explore hybrid optimization strategies and validate the framework against other bio-inspired algorithms to enhance its robustness and scalability.
Comparative Analysis of Image Enhancement Techniques for Brain Tumor Segmentation: Contrast, Histogram, and Hybrid Approaches
Apr 08, 2024
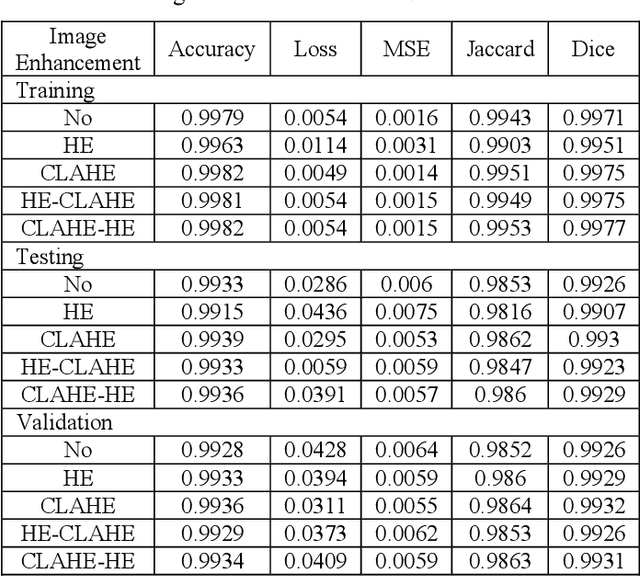

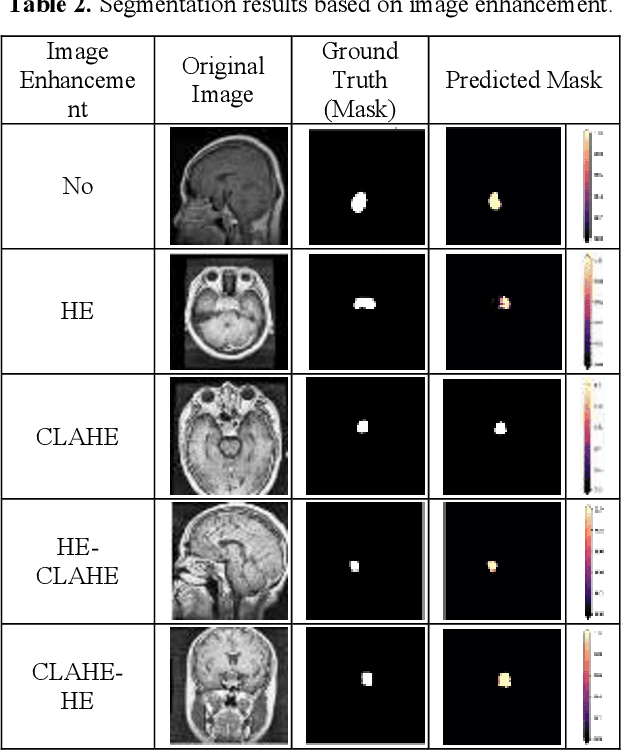
Abstract:This study systematically investigates the impact of image enhancement techniques on Convolutional Neural Network (CNN)-based Brain Tumor Segmentation, focusing on Histogram Equalization (HE), Contrast Limited Adaptive Histogram Equalization (CLAHE), and their hybrid variations. Employing the U-Net architecture on a dataset of 3064 Brain MRI images, the research delves into preprocessing steps, including resizing and enhancement, to optimize segmentation accuracy. A detailed analysis of the CNN-based U-Net architecture, training, and validation processes is provided. The comparative analysis, utilizing metrics such as Accuracy, Loss, MSE, IoU, and DSC, reveals that the hybrid approach CLAHE-HE consistently outperforms others. Results highlight its superior accuracy (0.9982, 0.9939, 0.9936 for training, testing, and validation, respectively) and robust segmentation overlap, with Jaccard values of 0.9862, 0.9847, and 0.9864, and Dice values of 0.993, 0.9923, and 0.9932 for the same phases, emphasizing its potential in neuro-oncological applications. The study concludes with a call for refinement in segmentation methodologies to further enhance diagnostic precision and treatment planning in neuro-oncology.
* 9 Pages, & Figures, 2 Tables, International Conference on Computer Science Electronics and Information (ICCSEI 2023)
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge