Pratima Upretee
Deep-learning assisted detection and quantification of (oo)cysts of Giardia and Cryptosporidium on smartphone microscopy images
Apr 11, 2023Abstract:The consumption of microbial-contaminated food and water is responsible for the deaths of millions of people annually. Smartphone-based microscopy systems are portable, low-cost, and more accessible alternatives for the detection of Giardia and Cryptosporidium than traditional brightfield microscopes. However, the images from smartphone microscopes are noisier and require manual cyst identification by trained technicians, usually unavailable in resource-limited settings. Automatic detection of (oo)cysts using deep-learning-based object detection could offer a solution for this limitation. We evaluate the performance of three state-of-the-art object detectors to detect (oo)cysts of Giardia and Cryptosporidium on a custom dataset that includes both smartphone and brightfield microscopic images from vegetable samples. Faster RCNN, RetinaNet, and you only look once (YOLOv8s) deep-learning models were employed to explore their efficacy and limitations. Our results show that while the deep-learning models perform better with the brightfield microscopy image dataset than the smartphone microscopy image dataset, the smartphone microscopy predictions are still comparable to the prediction performance of non-experts.
FixMatchSeg: Fixing FixMatch for Semi-Supervised Semantic Segmentation
Aug 02, 2022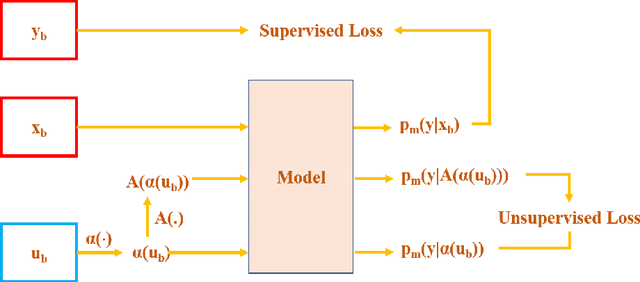
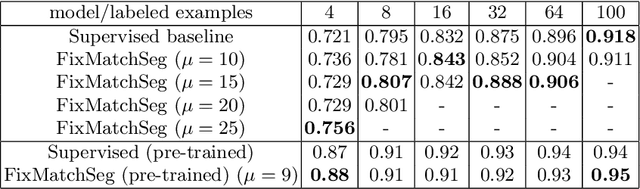
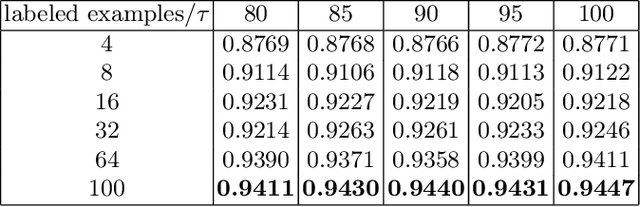
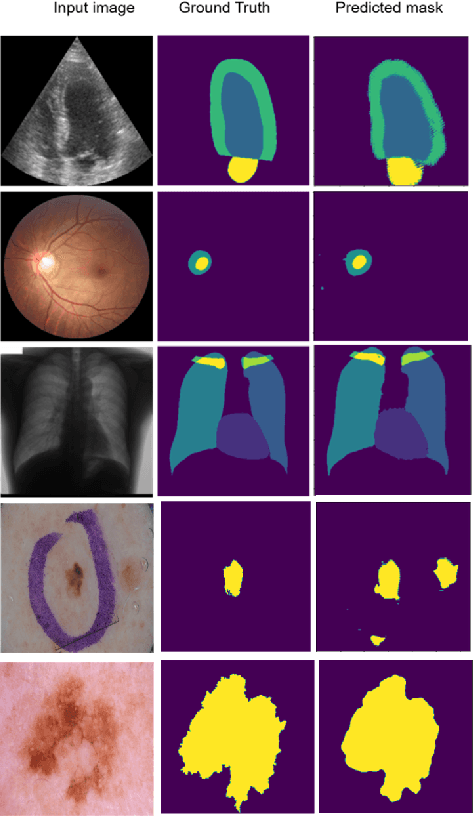
Abstract:Supervised deep learning methods for semantic medical image segmentation are getting increasingly popular in the past few years.However, in resource constrained settings, getting large number of annotated images is very difficult as it mostly requires experts, is expensive and time-consuming.Semi-supervised segmentation can be an attractive solution where a very few labeled images are used along with a large number of unlabeled ones. While the gap between supervised and semi-supervised methods have been dramatically reduced for classification problems in the past couple of years, there still remains a larger gap in segmentation methods. In this work, we adapt a state-of-the-art semi-supervised classification method FixMatch to semantic segmentation task, introducing FixMatchSeg. FixMatchSeg is evaluated in four different publicly available datasets of different anatomy and different modality: cardiac ultrasound, chest X-ray, retinal fundus image, and skin images. When there are few labels, we show that FixMatchSeg performs on par with strong supervised baselines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge