Pierre-Antoine Comby
MIND, JOLIOT
Robust plug-and-play methods for highly accelerated non-Cartesian MRI reconstruction
Nov 04, 2024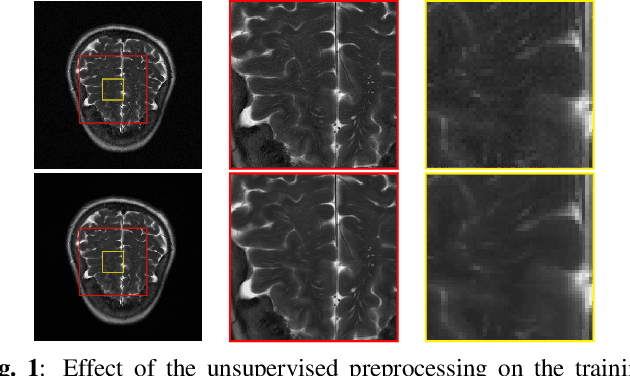
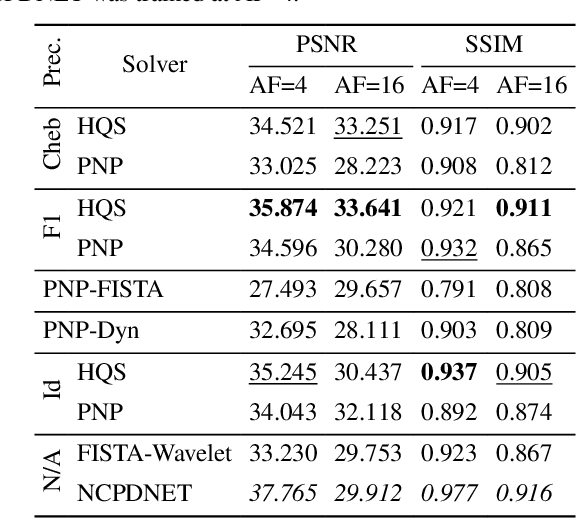
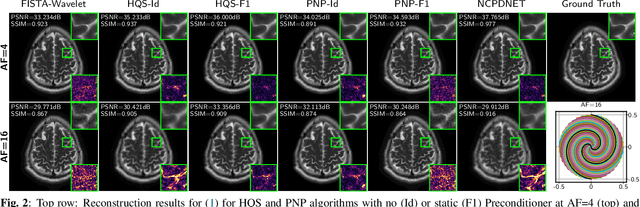
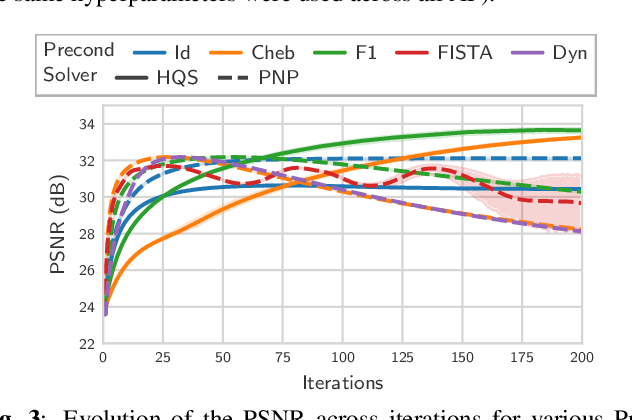
Abstract:Achieving high-quality Magnetic Resonance Imaging (MRI) reconstruction at accelerated acquisition rates remains challenging due to the inherent ill-posed nature of the inverse problem. Traditional Compressed Sensing (CS) methods, while robust across varying acquisition settings, struggle to maintain good reconstruction quality at high acceleration factors ($\ge$ 8). Recent advances in deep learning have improved reconstruction quality, but purely data-driven methods are prone to overfitting and hallucination effects, notably when the acquisition setting is varying. Plug-and-Play (PnP) approaches have been proposed to mitigate the pitfalls of both frameworks. In a nutshell, PnP algorithms amount to replacing suboptimal handcrafted CS priors with powerful denoising deep neural network (DNNs). However, in MRI reconstruction, existing PnP methods often yield suboptimal results due to instabilities in the proximal gradient descent (PGD) schemes and the lack of curated, noiseless datasets for training robust denoisers. In this work, we propose a fully unsupervised preprocessing pipeline to generate clean, noiseless complex MRI signals from multicoil data, enabling training of a high-performance denoising DNN. Furthermore, we introduce an annealed Half-Quadratic Splitting (HQS) algorithm to address the instability issues, leading to significant improvements over existing PnP algorithms. When combined with preconditioning techniques, our approach achieves state-of-the-art results, providing a robust and efficient solution for high-quality MRI reconstruction.
SNAKE-fMRI: A modular fMRI data simulator from the space-time domain to k-space and back
Apr 12, 2024Abstract:We propose a new, modular, open-source, Python-based 3D+time fMRI data simulation software, \emph{SNAKE-fMRI}, which stands for \emph{S}imulator from \emph{N}eurovascular coupling to \emph{A}cquisition of \emph{K}-space data for \emph{E}xploration of fMRI acquisition techniques.Unlike existing tools, the goal here is to simulate the complete chain of fMRI data acquisition, from the spatio-temporal design of evoked brain responses to various multi-coil k-space data 3D sampling strategies, with the possibility of extending the forward acquisition model to various noise and artifact sources while remaining memory-efficient.By using this \emph{in silico} setup, we are thus able to provide realistic and reproducible ground truth for fMRI reconstruction methods in 3D accelerated acquisition settings and explore the influence of critical parameters, such as the acceleration factor and signal-to-noise ratio~(SNR), on downstream tasks of image reconstruction and statistical analysis of evoked brain activity.We present three scenarios of increasing complexity to showcase the flexibility, versatility, and fidelity of \emph{SNAKE-fMRI}: From a temporally-fixed full 3D Cartesian to various 3D non-Cartesian sampling patterns, we can compare -- with reproducibility guarantees -- how experimental paradigms, acquisition strategies and reconstruction methods contribute and interact together, affecting the downstream statistical analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge