Pepijn Van de Ven
Image Segmentation: Inducing graph-based learning
Jan 07, 2025Abstract:This study explores the potential of graph neural networks (GNNs) to enhance semantic segmentation across diverse image modalities. We evaluate the effectiveness of a novel GNN-based U-Net architecture on three distinct datasets: PascalVOC, a standard benchmark for natural image segmentation, WoodScape, a challenging dataset of fisheye images commonly used in autonomous driving, introducing significant geometric distortions; and ISIC2016, a dataset of dermoscopic images for skin lesion segmentation. We compare our proposed UNet-GNN model against established convolutional neural networks (CNNs) based segmentation models, including U-Net and U-Net++, as well as the transformer-based SwinUNet. Unlike these methods, which primarily rely on local convolutional operations or global self-attention, GNNs explicitly model relationships between image regions by constructing and operating on a graph representation of the image features. This approach allows the model to capture long-range dependencies and complex spatial relationships, which we hypothesize will be particularly beneficial for handling geometric distortions present in fisheye imagery and capturing intricate boundaries in medical images. Our analysis demonstrates the versatility of GNNs in addressing diverse segmentation challenges and highlights their potential to improve segmentation accuracy in various applications, including autonomous driving and medical image analysis.
Subgraph Clustering and Atom Learning for Improved Image Classification
Jul 20, 2024Abstract:In this study, we present the Graph Sub-Graph Network (GSN), a novel hybrid image classification model merging the strengths of Convolutional Neural Networks (CNNs) for feature extraction and Graph Neural Networks (GNNs) for structural modeling. GSN employs k-means clustering to group graph nodes into clusters, facilitating the creation of subgraphs. These subgraphs are then utilized to learn representative `atoms` for dictionary learning, enabling the identification of sparse, class-distinguishable features. This integrated approach is particularly relevant in domains like medical imaging, where discerning subtle feature differences is crucial for accurate classification. To evaluate the performance of our proposed GSN, we conducted experiments on benchmark datasets, including PascalVOC and HAM10000. Our results demonstrate the efficacy of our model in optimizing dictionary configurations across varied classes, which contributes to its effectiveness in medical classification tasks. This performance enhancement is primarily attributed to the integration of CNNs, GNNs, and graph learning techniques, which collectively improve the handling of datasets with limited labeled examples. Specifically, our experiments show that the model achieves a higher accuracy on benchmark datasets such as Pascal VOC and HAM10000 compared to conventional CNN approaches.
Connecting the Dots: Graph Neural Network Powered Ensemble and Classification of Medical Images
Nov 13, 2023Abstract:Deep learning models have demonstrated remarkable results for various computer vision tasks, including the realm of medical imaging. However, their application in the medical domain is limited due to the requirement for large amounts of training data, which can be both challenging and expensive to obtain. To mitigate this, pre-trained models have been fine-tuned on domain-specific data, but such an approach can suffer from inductive biases. Furthermore, deep learning models struggle to learn the relationship between spatially distant features and their importance, as convolution operations treat all pixels equally. Pioneering a novel solution to this challenge, we employ the Image Foresting Transform to optimally segment images into superpixels. These superpixels are subsequently transformed into graph-structured data, enabling the proficient extraction of features and modeling of relationships using Graph Neural Networks (GNNs). Our method harnesses an ensemble of three distinct GNN architectures to boost its robustness. In our evaluations targeting pneumonia classification, our methodology surpassed prevailing Deep Neural Networks (DNNs) in performance, all while drastically cutting down on the parameter count. This not only trims down the expenses tied to data but also accelerates training and minimizes bias. Consequently, our proposition offers a sturdy, economically viable, and scalable strategy for medical image classification, significantly diminishing dependency on extensive training data sets.
* Our code is available at https://github.com/aryan-at-ul/AICS_2023_submission
Compact & Capable: Harnessing Graph Neural Networks and Edge Convolution for Medical Image Classification
Jul 24, 2023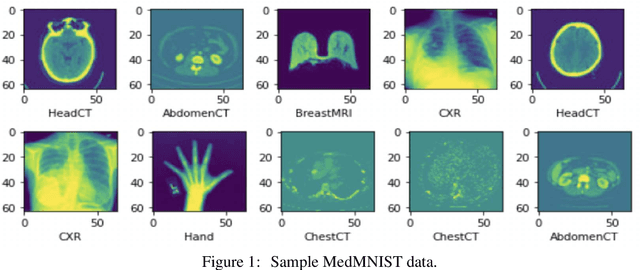
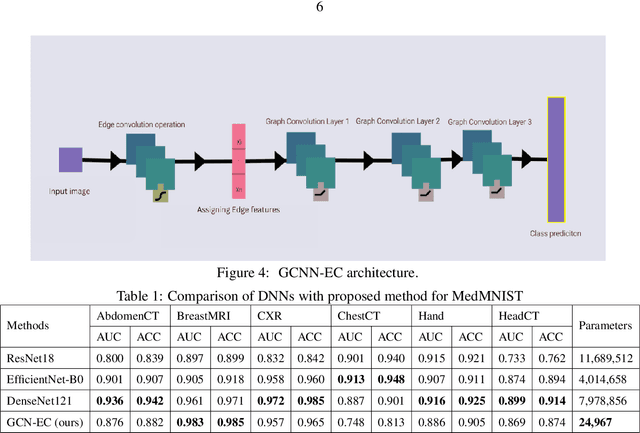
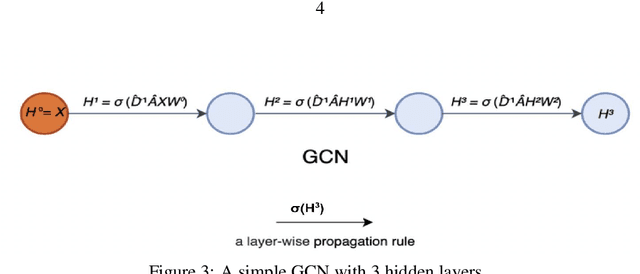
Abstract:Graph-based neural network models are gaining traction in the field of representation learning due to their ability to uncover latent topological relationships between entities that are otherwise challenging to identify. These models have been employed across a diverse range of domains, encompassing drug discovery, protein interactions, semantic segmentation, and fluid dynamics research. In this study, we investigate the potential of Graph Neural Networks (GNNs) for medical image classification. We introduce a novel model that combines GNNs and edge convolution, leveraging the interconnectedness of RGB channel feature values to strongly represent connections between crucial graph nodes. Our proposed model not only performs on par with state-of-the-art Deep Neural Networks (DNNs) but does so with 1000 times fewer parameters, resulting in reduced training time and data requirements. We compare our Graph Convolutional Neural Network (GCNN) to pre-trained DNNs for classifying MedMNIST dataset classes, revealing promising prospects for GNNs in medical image analysis. Our results also encourage further exploration of advanced graph-based models such as Graph Attention Networks (GAT) and Graph Auto-Encoders in the medical imaging domain. The proposed model yields more reliable, interpretable, and accurate outcomes for tasks like semantic segmentation and image classification compared to simpler GCNNs
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge