Paweł Russek
Computer-Aided Cytology Diagnosis in Animals: CNN-Based Image Quality Assessment for Accurate Disease Classification
Aug 11, 2023Abstract:This paper presents a computer-aided cytology diagnosis system designed for animals, focusing on image quality assessment (IQA) using Convolutional Neural Networks (CNNs). The system's building blocks are tailored to seamlessly integrate IQA, ensuring reliable performance in disease classification. We extensively investigate the CNN's ability to handle various image variations and scenarios, analyzing the impact on detecting low-quality input data. Additionally, the network's capacity to differentiate valid cellular samples from those with artifacts is evaluated. Our study employs a ResNet18 network architecture and explores the effects of input sizes and cropping strategies on model performance. The research sheds light on the significance of CNN-based IQA in computer-aided cytology diagnosis for animals, enhancing the accuracy of disease classification.
Using simulation to calibrate real data acquisition in veterinary medicine
Jul 21, 2023Abstract:This paper explores the innovative use of simulation environments to enhance data acquisition and diagnostics in veterinary medicine, focusing specifically on gait analysis in dogs. The study harnesses the power of Blender and the Blenderproc library to generate synthetic datasets that reflect diverse anatomical, environmental, and behavioral conditions. The generated data, represented in graph form and standardized for optimal analysis, is utilized to train machine learning algorithms for identifying normal and abnormal gaits. Two distinct datasets with varying degrees of camera angle granularity are created to further investigate the influence of camera perspective on model accuracy. Preliminary results suggest that this simulation-based approach holds promise for advancing veterinary diagnostics by enabling more precise data acquisition and more effective machine learning models. By integrating synthetic and real-world patient data, the study lays a robust foundation for improving overall effectiveness and efficiency in veterinary medicine.
Using super-resolution for enhancing visual perception and segmentation performance in veterinary cytology
Jun 20, 2023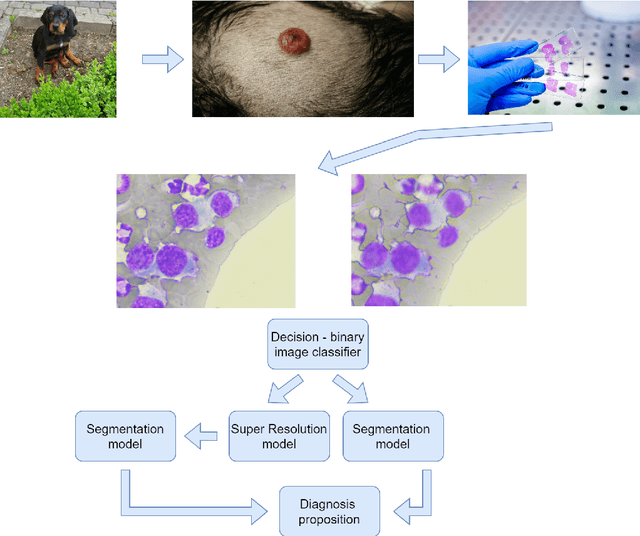
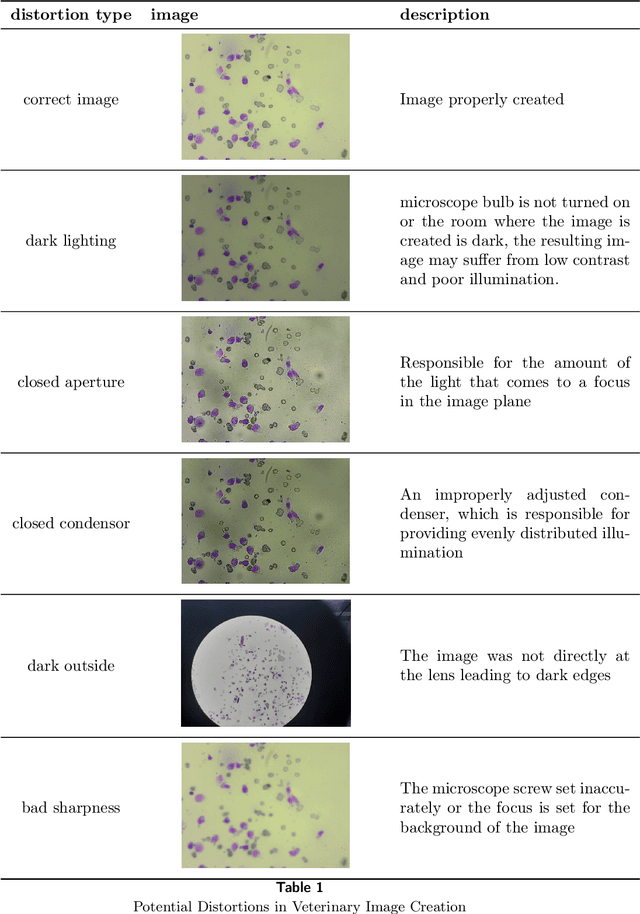
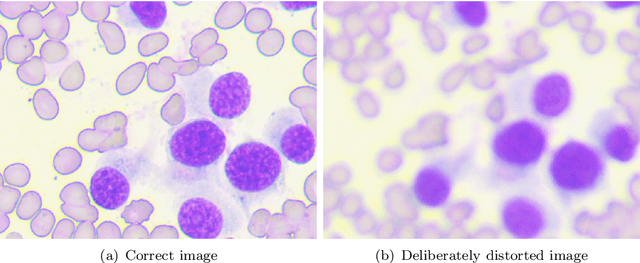
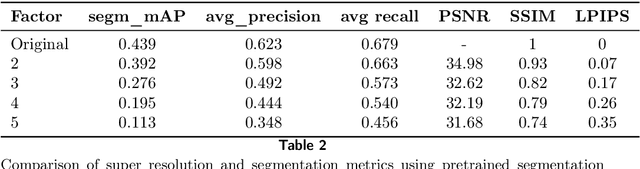
Abstract:The primary objective of this research was to enhance the quality of semantic segmentation in cytology images by incorporating super-resolution (SR) architectures. An additional contribution was the development of a novel dataset aimed at improving imaging quality in the presence of inaccurate focus. Our experimental results demonstrate that the integration of SR techniques into the segmentation pipeline can lead to a significant improvement of up to 25% in the mean average precision (mAP) segmentation metric. These findings suggest that leveraging SR architectures holds great promise for advancing the state of the art in cytology image analysis.
Segmentation of the veterinary cytological images for fast neoplastic tumors diagnosis
May 07, 2023



Abstract:This paper shows the machine learning system which performs instance segmentation of cytological images in veterinary medicine. Eleven cell types were used directly and indirectly in the experiments, including damaged and unrecognized categories. The deep learning models employed in the system achieve a high score of average precision and recall metrics, i.e. 0.94 and 0.8 respectively, for the selected three types of tumors. This variety of label types allowed us to draw a meaningful conclusion that there are relatively few mistakes for tumor cell types. Additionally, the model learned tumor cell features well enough to avoid misclassification mistakes of one tumor type into another. The experiments also revealed that the quality of the results improves with the dataset size (excluding the damaged cells). It is worth noting that all the experiments were done using a custom dedicated dataset provided by the cooperating vet doctors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge