Oğuz Dikenelli
Relation Extraction Capabilities of LLMs on Clinical Text: A Bilingual Evaluation for English and Turkish
Jan 14, 2026Abstract:The scarcity of annotated datasets for clinical information extraction in non-English languages hinders the evaluation of large language model (LLM)-based methods developed primarily in English. In this study, we present the first comprehensive bilingual evaluation of LLMs for the clinical Relation Extraction (RE) task in both English and Turkish. To facilitate this evaluation, we introduce the first English-Turkish parallel clinical RE dataset, derived and carefully curated from the 2010 i2b2/VA relation classification corpus. We systematically assess a diverse set of prompting strategies, including multiple in-context learning (ICL) and Chain-of-Thought (CoT) approaches, and compare their performance to fine-tuned baselines such as PURE. Furthermore, we propose Relation-Aware Retrieval (RAR), a novel in-context example selection method based on contrastive learning, that is specifically designed to capture both sentence-level and relation-level semantics. Our results show that prompting-based LLM approaches consistently outperform traditional fine-tuned models. Moreover, evaluations for English performed better than their Turkish counterparts across all evaluated LLMs and prompting techniques. Among ICL methods, RAR achieves the highest performance, with Gemini 1.5 Flash reaching a micro-F1 score of 0.906 in English and 0.888 in Turkish. Performance further improves to 0.918 F1 in English when RAR is combined with a structured reasoning prompt using the DeepSeek-V3 model. These findings highlight the importance of high-quality demonstration retrieval and underscore the potential of advanced retrieval and prompting techniques to bridge resource gaps in clinical natural language processing.
Harnessing the Power of BERT in the Turkish Clinical Domain: Pretraining Approaches for Limited Data Scenarios
May 05, 2023Abstract:In recent years, major advancements in natural language processing (NLP) have been driven by the emergence of large language models (LLMs), which have significantly revolutionized research and development within the field. Building upon this progress, our study delves into the effects of various pre-training methodologies on Turkish clinical language models' performance in a multi-label classification task involving radiology reports, with a focus on addressing the challenges posed by limited language resources. Additionally, we evaluated the simultaneous pretraining approach by utilizing limited clinical task data for the first time. We developed four models, including TurkRadBERT-task v1, TurkRadBERT-task v2, TurkRadBERT-sim v1, and TurkRadBERT-sim v2. Our findings indicate that the general Turkish BERT model (BERTurk) and TurkRadBERT-task v1, both of which utilize knowledge from a substantial general-domain corpus, demonstrate the best overall performance. Although the task-adaptive pre-training approach has the potential to capture domain-specific patterns, it is constrained by the limited task-specific corpus and may be susceptible to overfitting. Furthermore, our results underscore the significance of domain-specific vocabulary during pre-training for enhancing model performance. Ultimately, we observe that the combination of general-domain knowledge and task-specific fine-tuning is essential for achieving optimal performance across a range of categories. This study offers valuable insights for developing effective Turkish clinical language models and can guide future research on pre-training techniques for other low-resource languages within the clinical domain.
Attention-Wrapped Hierarchical BLSTMs for DDI Extraction
Jul 31, 2019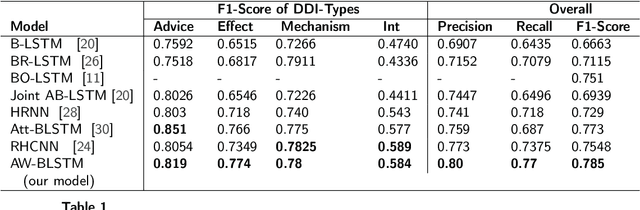
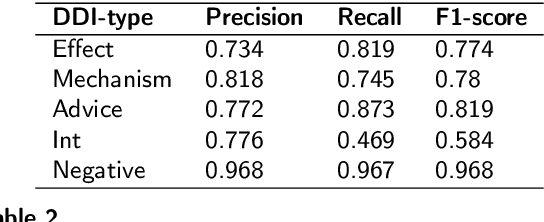
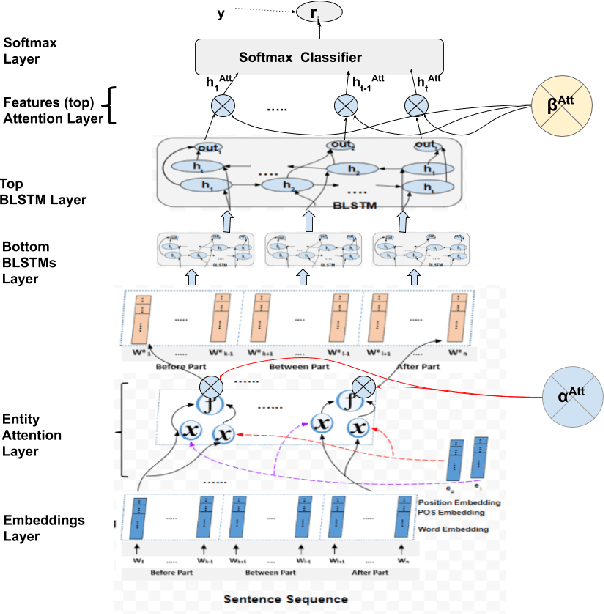
Abstract:Drug-Drug Interactions (DDIs) Extraction refers to the efforts to generate hand-made or automatic tools to extract embedded information from text and literature in the biomedical domain. Because of restrictions in hand-made efforts and their lower speed, Machine-Learning, or Deep-Learning approaches have become more popular for extracting DDIs. In this study, we propose a novel and generic Deep-Learning model which wraps Hierarchical Bidirectional LSTMs with two Attention Mechanisms that outperforms state-of-the-art models for DDIs Extraction, based on the DDIExtraction-2013 corpora. This model has obtained the macro F1-score of 0.785, and the precision of 0.80.
Agent-based Exploration of Wirings of Biological Neural Networks: Position Paper
Sep 14, 2012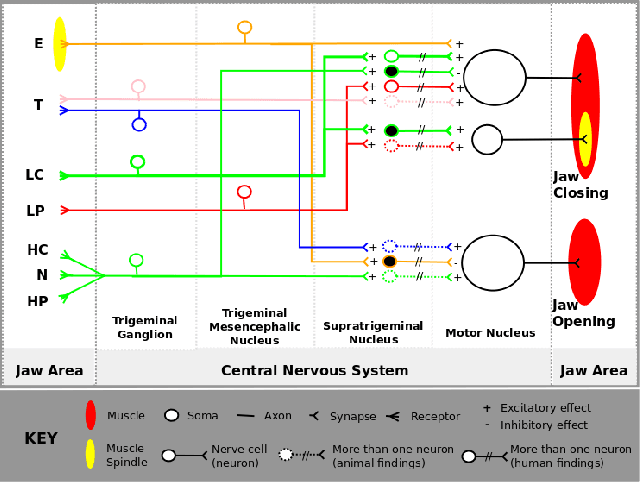
Abstract:The understanding of human central nervous system depends on knowledge of its wiring. However, there are still gaps in our understanding of its wiring due to technical difficulties. While some information is coming out from human experiments, medical research is lacking of simulation models to put current findings together to obtain the global picture and to predict hypotheses to lead future experiments. Agent-based modeling and simulation (ABMS) is a strong candidate for the simulation model. In this position paper, we discuss the current status of "neural wiring" and "ABMS in biological systems". In particular, we discuss that the ABMS context provides features required for exploration of biological neural wiring.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge