Noor Nakhaei
Spatial Matching of 2D Mammography Images and Specimen Radiographs: Towards Improved Characterization of Suspicious Microcalcifications
May 21, 2024Abstract:Accurate characterization of suspicious microcalcifications is critical to determine whether these calcifications are associated with invasive disease. Our overarching objective is to enable the joint characterization of microcalcifications and surrounding breast tissue using mammography images and digital histopathology images. Towards this goal, we investigate a template matching-based approach that utilizes microcalcifications as landmarks to match radiographs taken of biopsy core specimens to groups of calcifications that are visible on mammography. Our approach achieved a high negative predictive value (0.98) but modest precision (0.66) and recall (0.58) in identifying the mammographic region where microcalcifications were taken during a core needle biopsy.
Segmentation of Breast Microcalcifications: A Multi-Scale Approach
Feb 01, 2021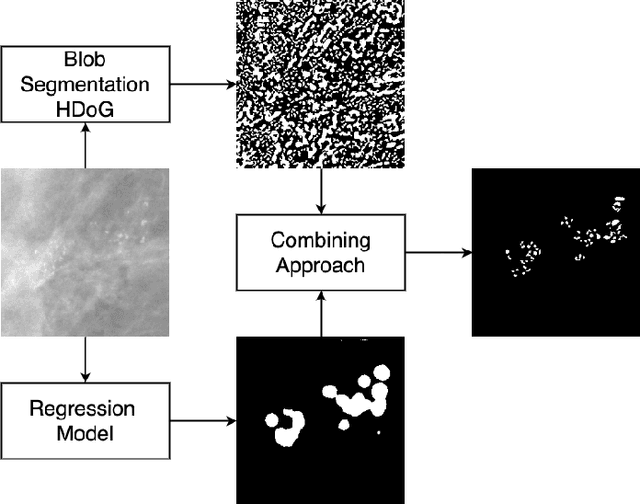
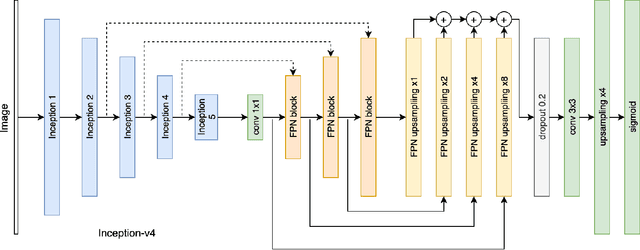
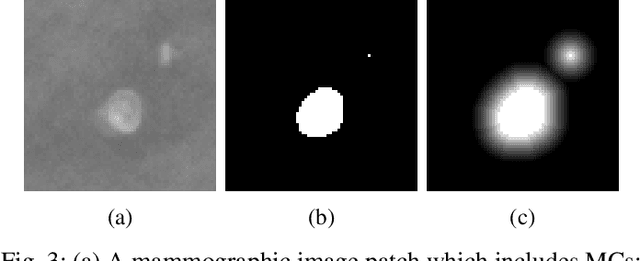
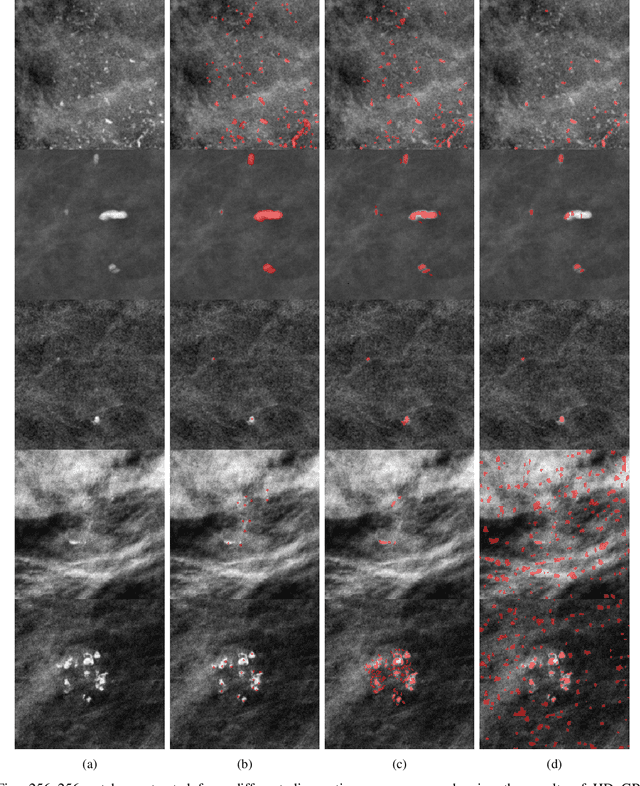
Abstract:Accurate characterization of microcalcifications (MCs) in 2D full-field digital screening mammography is a necessary step towards reducing diagnostic uncertainty associated with the callback of women with suspicious MCs. Quantitative analysis of MCs has the potential to better identify MCs that have a higher likelihood of corresponding to invasive cancer. However, automated identification and segmentation of MCs remains a challenging task with high false positive rates. We present Hessian Difference of Gaussians Regression (HDoGReg), a two stage multi-scale approach to MC segmentation. Candidate high optical density objects are first delineated using blob detection and Hessian analysis. A regression convolutional network, trained to output a function with higher response near MCs, chooses the objects which constitute actual MCs. The method is trained and validated on 435 mammograms from two separate datasets. HDoGReg achieved a mean intersection over the union of 0.670$\pm$0.121 per image, intersection over the union per MC object of 0.607$\pm$0.250 and true positive rate of 0.744 at 0.4 false positive detections per $cm^2$. The results of HDoGReg perform better when compared to state-of-the-art MC segmentation and detection methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge