Nicola Amoroso
Deep convolutional framelets for dose reconstruction in BNCT with Compton camera detector
Sep 24, 2024Abstract:Boron Neutron Capture Therapy (BNCT) is an innovative binary form of radiation therapy with high selectivity towards cancer tissue based on the neutron capture reaction 10B(n,$\alpha$)7Li, consisting in the exposition of patients to neutron beams after administration of a boron compound with preferential accumulation in cancer cells. The high linear energy transfer products of the ensuing reaction deposit their energy at cell level, sparing normal tissue. Although progress in accelerator-based BNCT has led to renewed interest in this cancer treatment modality, in vivo dose monitoring during treatment still remains not feasible and several approaches are under investigation. While Compton imaging presents various advantages over other imaging methods, it typically requires long reconstruction times, comparable with BNCT treatment duration. This study aims to develop deep neural network models to estimate the dose distribution by using a simulated dataset of BNCT Compton camera images. The models pursue the avoidance of the iteration time associated with the maximum-likelihood expectation-maximization algorithm (MLEM), enabling a prompt dose reconstruction during the treatment. The U-Net architecture and two variants based on the deep convolutional framelets framework have been used for noise and artifacts reduction in few-iterations reconstructed images, leading to promising results in terms of reconstruction accuracy and processing time.
Feature Selection based on Machine Learning in MRIs for Hippocampal Segmentation
Jan 16, 2015
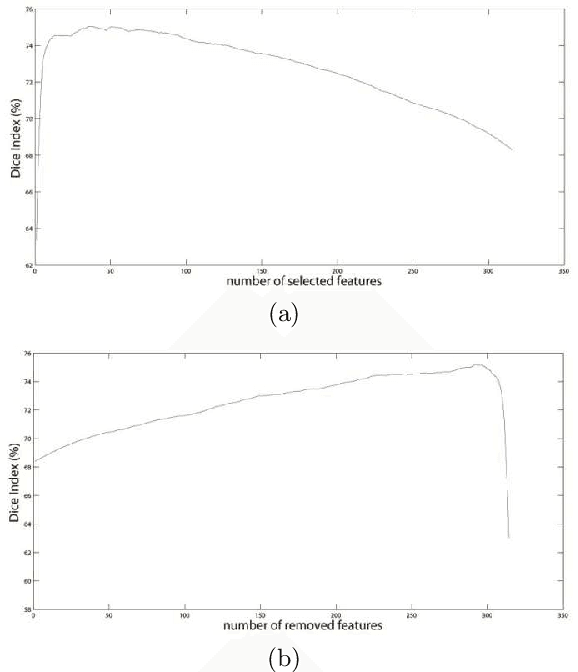

Abstract:Neurodegenerative diseases are frequently associated with structural changes in the brain. Magnetic Resonance Imaging (MRI) scans can show these variations and therefore be used as a supportive feature for a number of neurodegenerative diseases. The hippocampus has been known to be a biomarker for Alzheimer disease and other neurological and psychiatric diseases. However, it requires accurate, robust and reproducible delineation of hippocampal structures. Fully automatic methods are usually the voxel based approach, for each voxel a number of local features were calculated. In this paper we compared four different techniques for feature selection from a set of 315 features extracted for each voxel: (i) filter method based on the Kolmogorov-Smirnov test; two wrapper methods, respectively, (ii) Sequential Forward Selection and (iii) Sequential Backward Elimination; and (iv) embedded method based on the Random Forest Classifier on a set of 10 T1-weighted brain MRIs and tested on an independent set of 25 subjects. The resulting segmentations were compared with manual reference labelling. By using only 23 features for each voxel (sequential backward elimination) we obtained comparable state of-the-art performances with respect to the standard tool FreeSurfer.
* To appear on "Computational and Mathematical Methods in Medicine", Hindawi Publishing Corporation. 19 pages, 7 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge