Nicholas G. Reich
Flusion: Integrating multiple data sources for accurate influenza predictions
Jul 26, 2024Abstract:Over the last ten years, the US Centers for Disease Control and Prevention (CDC) has organized an annual influenza forecasting challenge with the motivation that accurate probabilistic forecasts could improve situational awareness and yield more effective public health actions. Starting with the 2021/22 influenza season, the forecasting targets for this challenge have been based on hospital admissions reported in the CDC's National Healthcare Safety Network (NHSN) surveillance system. Reporting of influenza hospital admissions through NHSN began within the last few years, and as such only a limited amount of historical data are available for this signal. To produce forecasts in the presence of limited data for the target surveillance system, we augmented these data with two signals that have a longer historical record: 1) ILI+, which estimates the proportion of outpatient doctor visits where the patient has influenza; and 2) rates of laboratory-confirmed influenza hospitalizations at a selected set of healthcare facilities. Our model, Flusion, is an ensemble that combines gradient boosting quantile regression models with a Bayesian autoregressive model. The gradient boosting models were trained on all three data signals, while the autoregressive model was trained on only the target signal; all models were trained jointly on data for multiple locations. Flusion was the top-performing model in the CDC's influenza prediction challenge for the 2023/24 season. In this article we investigate the factors contributing to Flusion's success, and we find that its strong performance was primarily driven by the use of a gradient boosting model that was trained jointly on data from multiple surveillance signals and locations. These results indicate the value of sharing information across locations and surveillance signals, especially when doing so adds to the pool of available training data.
Adaptively stacking ensembles for influenza forecasting with incomplete data
Jul 26, 2019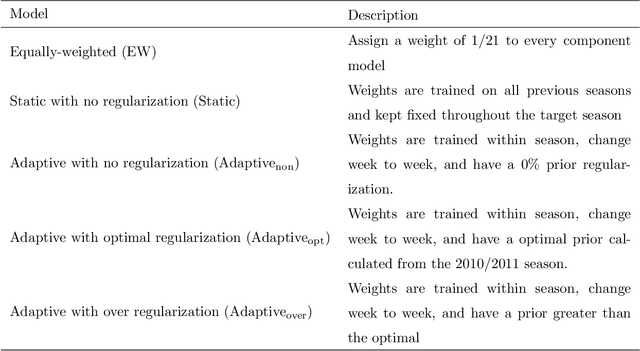
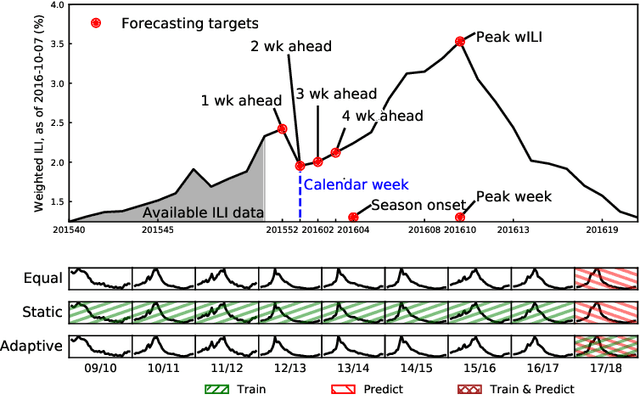
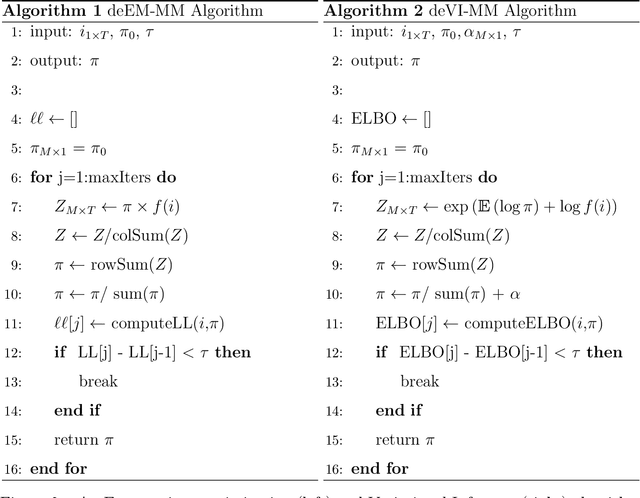
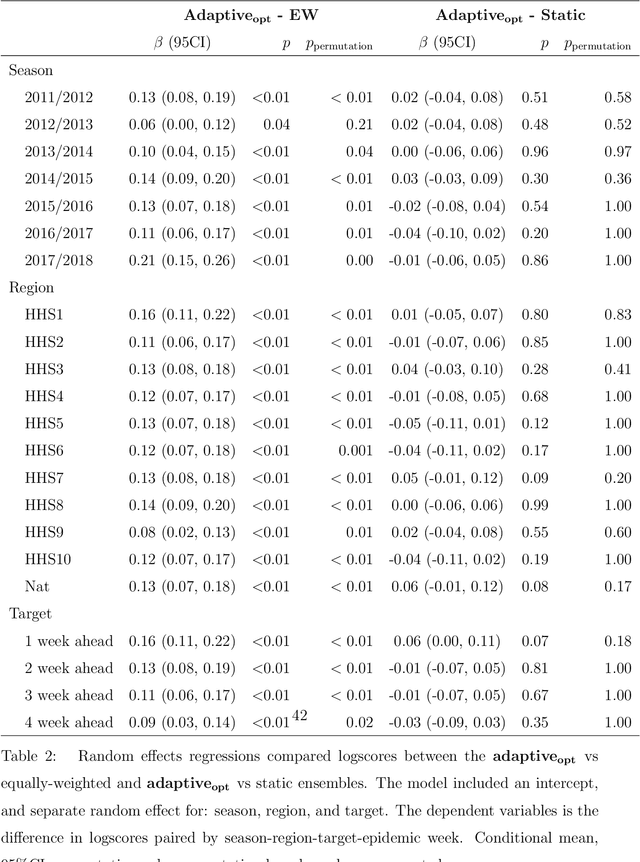
Abstract:Seasonal influenza infects between 10 and 50 million people in the United States every year, overburdening hospitals during weeks of peak incidence. Named by the CDC as an important tool to fight the damaging effects of these epidemics, accurate forecasts of influenza and influenza-like illness (ILI) forewarn public health officials about when, and where, seasonal influenza outbreaks will hit hardest. Multi-model ensemble forecasts---weighted combinations of component models---have shown positive results in forecasting. Ensemble forecasts of influenza outbreaks have been static, training on all past ILI data at the beginning of a season, generating a set of optimal weights for each model in the ensemble, and keeping the weights constant. We propose an adaptive ensemble forecast that (i) changes model weights week-by-week throughout the influenza season, (ii) only needs the current influenza season's data to make predictions, and (iii) by introducing a prior distribution, shrinks weights toward the reference equal weighting approach and adjusts for observed ILI percentages that are subject to future revisions. We investigate the prior's ability to impact adaptive ensemble performance and, after finding an optimal prior via a cross-validation approach, compare our adaptive ensemble's performance to equal-weighted and static ensembles. Applied to forecasts of short-term ILI incidence at the regional and national level in the US, our adaptive model outperforms a naive equal-weighted ensemble, and has similar or better performance to the static ensemble, which requires multiple years of training data. Adaptive ensembles are able to quickly train and forecast during epidemics, and provide a practical tool to public health officials looking for forecasts that can conform to unique features of a specific season.
Prediction of infectious disease epidemics via weighted density ensembles
Mar 31, 2017



Abstract:Accurate and reliable predictions of infectious disease dynamics can be valuable to public health organizations that plan interventions to decrease or prevent disease transmission. A great variety of models have been developed for this task, using different model structures, covariates, and targets for prediction. Experience has shown that the performance of these models varies; some tend to do better or worse in different seasons or at different points within a season. Ensemble methods combine multiple models to obtain a single prediction that leverages the strengths of each model. We considered a range of ensemble methods that each form a predictive density for a target of interest as a weighted sum of the predictive densities from component models. In the simplest case, equal weight is assigned to each component model; in the most complex case, the weights vary with the region, prediction target, week of the season when the predictions are made, a measure of component model uncertainty, and recent observations of disease incidence. We applied these methods to predict measures of influenza season timing and severity in the United States, both at the national and regional levels, using three component models. We trained the models on retrospective predictions from 14 seasons (1997/1998 - 2010/2011) and evaluated each model's prospective, out-of-sample performance in the five subsequent influenza seasons. In this test phase, the ensemble methods showed overall performance that was similar to the best of the component models, but offered more consistent performance across seasons than the component models. Ensemble methods offer the potential to deliver more reliable predictions to public health decision makers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge