Nguyen Thi Oanh
UGCANet: A Unified Global Context-Aware Transformer-based Network with Feature Alignment for Endoscopic Image Analysis
Jul 12, 2023Abstract:Gastrointestinal endoscopy is a medical procedure that utilizes a flexible tube equipped with a camera and other instruments to examine the digestive tract. This minimally invasive technique allows for diagnosing and managing various gastrointestinal conditions, including inflammatory bowel disease, gastrointestinal bleeding, and colon cancer. The early detection and identification of lesions in the upper gastrointestinal tract and the identification of malignant polyps that may pose a risk of cancer development are critical components of gastrointestinal endoscopy's diagnostic and therapeutic applications. Therefore, enhancing the detection rates of gastrointestinal disorders can significantly improve a patient's prognosis by increasing the likelihood of timely medical intervention, which may prolong the patient's lifespan and improve overall health outcomes. This paper presents a novel Transformer-based deep neural network designed to perform multiple tasks simultaneously, thereby enabling accurate identification of both upper gastrointestinal tract lesions and colon polyps. Our approach proposes a unique global context-aware module and leverages the powerful MiT backbone, along with a feature alignment block, to enhance the network's representation capability. This novel design leads to a significant improvement in performance across various endoscopic diagnosis tasks. Extensive experiments demonstrate the superior performance of our method compared to other state-of-the-art approaches.
RaBiT: An Efficient Transformer using Bidirectional Feature Pyramid Network with Reverse Attention for Colon Polyp Segmentation
Jul 12, 2023



Abstract:Automatic and accurate segmentation of colon polyps is essential for early diagnosis of colorectal cancer. Advanced deep learning models have shown promising results in polyp segmentation. However, they still have limitations in representing multi-scale features and generalization capability. To address these issues, this paper introduces RaBiT, an encoder-decoder model that incorporates a lightweight Transformer-based architecture in the encoder to model multiple-level global semantic relationships. The decoder consists of several bidirectional feature pyramid layers with reverse attention modules to better fuse feature maps at various levels and incrementally refine polyp boundaries. We also propose ideas to lighten the reverse attention module and make it more suitable for multi-class segmentation. Extensive experiments on several benchmark datasets show that our method outperforms existing methods across all datasets while maintaining low computational complexity. Moreover, our method demonstrates high generalization capability in cross-dataset experiments, even when the training and test sets have different characteristics.
ColonFormer: An Efficient Transformer based Method for Colon Polyp Segmentation
May 17, 2022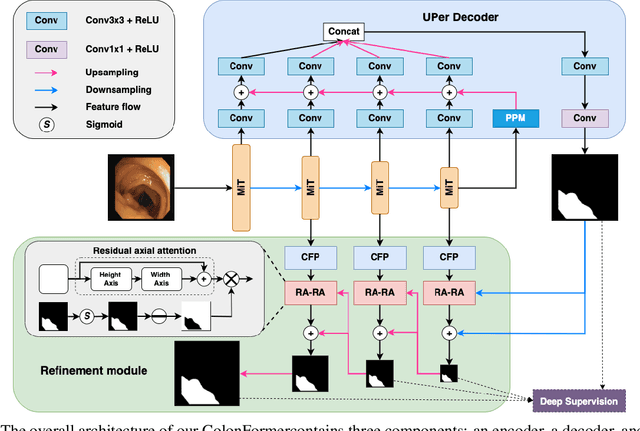
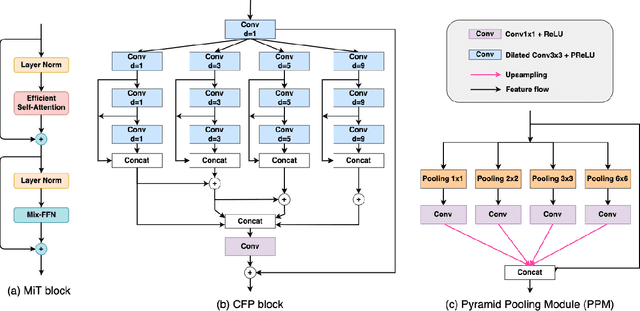
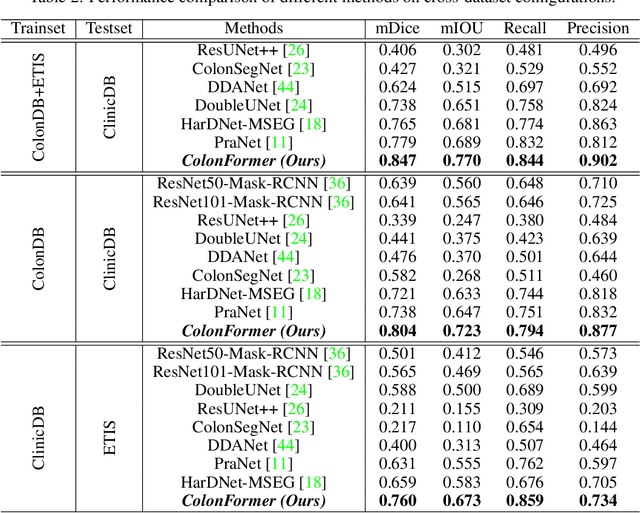
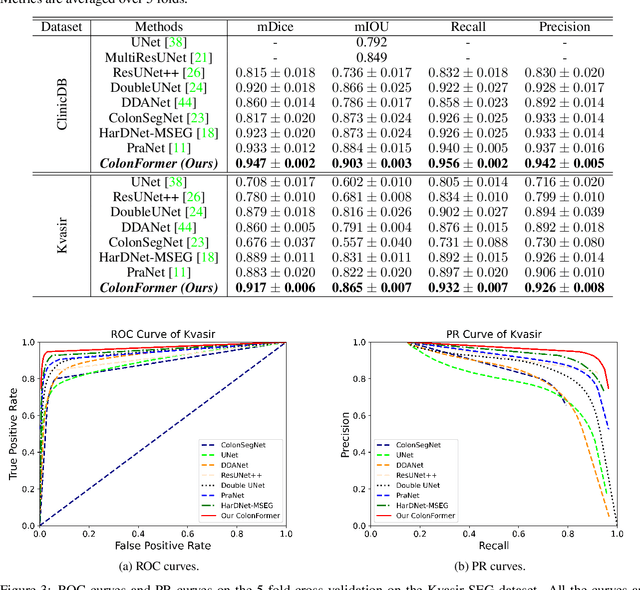
Abstract:Identifying polyps is a challenging problem for automatic analysis of endoscopic images in computer-aided clinical support systems. Models based on convolutional networks (CNN), transformers, and combinations of them have been proposed to segment polyps with promising results. However, those approaches have limitations either in modeling the local appearance of the polyps only or lack of multi-level features for spatial dependency in the decoding process. This paper proposes a novel network, namely ColonFormer, to address these limitations. ColonFormer is an encoder-decoder architecture with the capability of modeling long-range semantic information at both encoder and decoder branches. The encoder is a lightweight architecture based on transformers for modeling global semantic relations at multi scales. The decoder is a hierarchical network structure designed for learning multi-level features to enrich feature representation. Besides, a refinement module is added with a new skip connection technique to refine the boundary of polyp objects in the global map for accurate segmentation. Extensive experiments have been conducted on five popular benchmark datasets for polyp segmentation, including Kvasir, CVC-Clinic DB, CVCColonDB, EndoScene, and ETIS. Experimental results show that our ColonFormer achieve state-of-the-art performance on all benchmark datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge