Nguyen Thanh Duc
CV-Attention UNet: Attention-based UNet for 3D Cerebrovascular Segmentation of Enhanced TOF-MRA Images
Nov 16, 2023



Abstract:Due to the lack of automated methods, to diagnose cerebrovascular disease, time-of-flight magnetic resonance angiography (TOF-MRA) is assessed visually, making it time-consuming. The commonly used encoder-decoder architectures for cerebrovascular segmentation utilize redundant features, eventually leading to the extraction of low-level features multiple times. Additionally, convolutional neural networks (CNNs) suffer from performance degradation when the batch size is small, and deeper networks experience the vanishing gradient problem. Methods: In this paper, we attempt to solve these limitations and propose the 3D cerebrovascular attention UNet method, named CV-AttentionUNet, for precise extraction of brain vessel images. We proposed a sequence of preprocessing techniques followed by deeply supervised UNet to improve the accuracy of segmentation of the brain vessels leading to a stroke. To combine the low and high semantics, we applied the attention mechanism. This mechanism focuses on relevant associations and neglects irrelevant anatomical information. Furthermore, the inclusion of deep supervision incorporates different levels of features that prove to be beneficial for network convergence. Results: We demonstrate the efficiency of the proposed method by cross-validating with an unlabeled dataset, which was further labeled by us. We believe that the novelty of this algorithm lies in its ability to perform well on both labeled and unlabeled data with image processing-based enhancement. The results indicate that our method performed better than the existing state-of-the-art methods on the TubeTK dataset. Conclusion: The proposed method will help in accurate segmentation of cerebrovascular structure leading to stroke
ColonFormer: An Efficient Transformer based Method for Colon Polyp Segmentation
May 17, 2022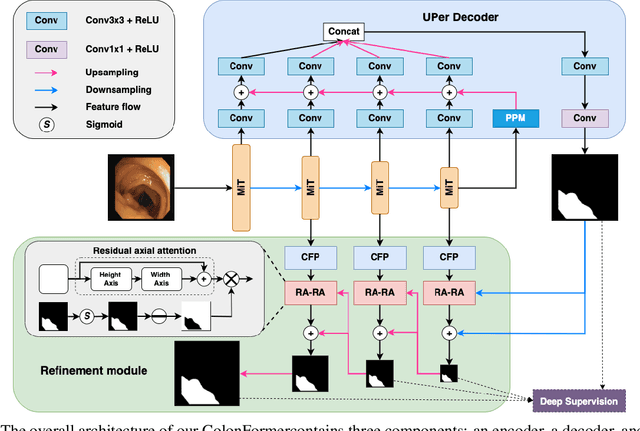
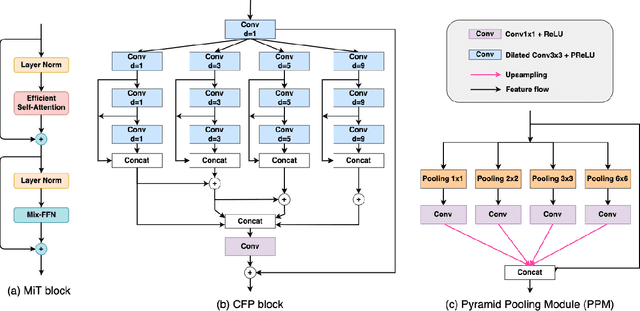
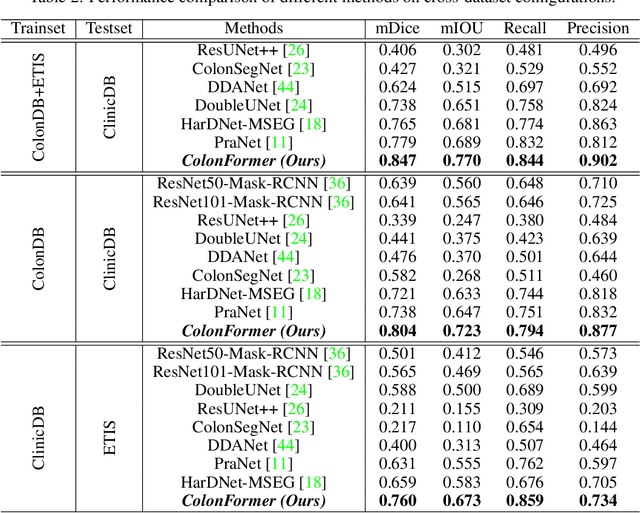
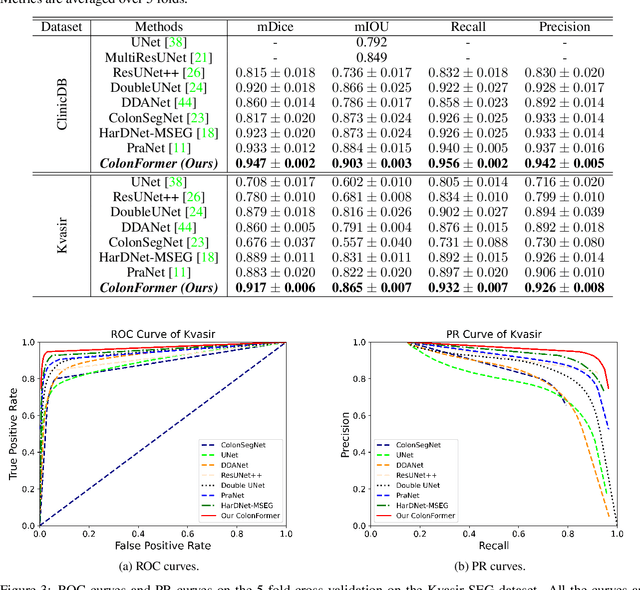
Abstract:Identifying polyps is a challenging problem for automatic analysis of endoscopic images in computer-aided clinical support systems. Models based on convolutional networks (CNN), transformers, and combinations of them have been proposed to segment polyps with promising results. However, those approaches have limitations either in modeling the local appearance of the polyps only or lack of multi-level features for spatial dependency in the decoding process. This paper proposes a novel network, namely ColonFormer, to address these limitations. ColonFormer is an encoder-decoder architecture with the capability of modeling long-range semantic information at both encoder and decoder branches. The encoder is a lightweight architecture based on transformers for modeling global semantic relations at multi scales. The decoder is a hierarchical network structure designed for learning multi-level features to enrich feature representation. Besides, a refinement module is added with a new skip connection technique to refine the boundary of polyp objects in the global map for accurate segmentation. Extensive experiments have been conducted on five popular benchmark datasets for polyp segmentation, including Kvasir, CVC-Clinic DB, CVCColonDB, EndoScene, and ETIS. Experimental results show that our ColonFormer achieve state-of-the-art performance on all benchmark datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge