Navneet Singh
RSNA Large Language Model Benchmark Dataset for Chest Radiographs of Cardiothoracic Disease: Radiologist Evaluation and Validation Enhanced by AI Labels (REVEAL-CXR)
Jan 21, 2026Abstract:Multimodal large language models have demonstrated comparable performance to that of radiology trainees on multiple-choice board-style exams. However, to develop clinically useful multimodal LLM tools, high-quality benchmarks curated by domain experts are essential. To curate released and holdout datasets of 100 chest radiographic studies each and propose an artificial intelligence (AI)-assisted expert labeling procedure to allow radiologists to label studies more efficiently. A total of 13,735 deidentified chest radiographs and their corresponding reports from the MIDRC were used. GPT-4o extracted abnormal findings from the reports, which were then mapped to 12 benchmark labels with a locally hosted LLM (Phi-4-Reasoning). From these studies, 1,000 were sampled on the basis of the AI-suggested benchmark labels for expert review; the sampling algorithm ensured that the selected studies were clinically relevant and captured a range of difficulty levels. Seventeen chest radiologists participated, and they marked "Agree all", "Agree mostly" or "Disagree" to indicate their assessment of the correctness of the LLM suggested labels. Each chest radiograph was evaluated by three experts. Of these, at least two radiologists selected "Agree All" for 381 radiographs. From this set, 200 were selected, prioritizing those with less common or multiple finding labels, and divided into 100 released radiographs and 100 reserved as the holdout dataset. The holdout dataset is used exclusively by RSNA to independently evaluate different models. A benchmark of 200 chest radiographic studies with 12 benchmark labels was created and made publicly available https://imaging.rsna.org, with each chest radiograph verified by three radiologists. In addition, an AI-assisted labeling procedure was developed to help radiologists label at scale, minimize unnecessary omissions, and support a semicollaborative environment.
Modeling Quantum Machine Learning for Genomic Data Analysis
Jan 14, 2025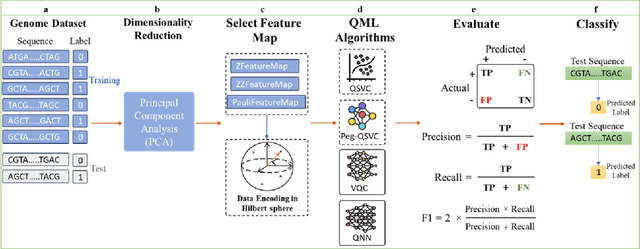
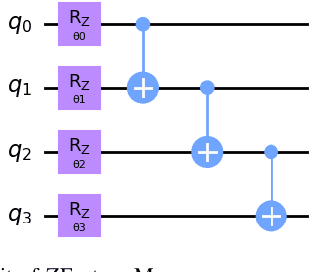
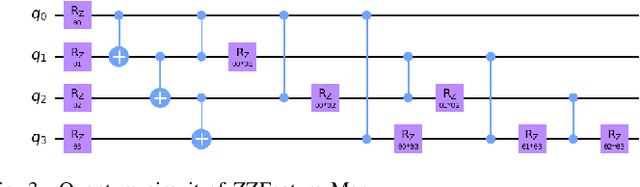
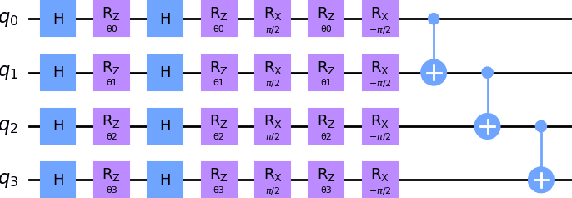
Abstract:Quantum Machine Learning (QML) continues to evolve, unlocking new opportunities for diverse applications. In this study, we investigate and evaluate the applicability of QML models for binary classification of genome sequence data by employing various feature mapping techniques. We present an open-source, independent Qiskit-based implementation to conduct experiments on a benchmark genomic dataset. Our simulations reveal that the interplay between feature mapping techniques and QML algorithms significantly influences performance. Notably, the Pegasos Quantum Support Vector Classifier (Pegasos-QSVC) exhibits high sensitivity, particularly excelling in recall metrics, while Quantum Neural Networks (QNN) achieve the highest training accuracy across all feature maps. However, the pronounced variability in classifier performance, dependent on feature mapping, highlights the risk of overfitting to localized output distributions in certain scenarios. This work underscores the transformative potential of QML for genomic data classification while emphasizing the need for continued advancements to enhance the robustness and accuracy of these methodologies.
Modeling Feature Maps for Quantum Machine Learning
Jan 14, 2025Abstract:Quantum Machine Learning (QML) offers significant potential for complex tasks like genome sequence classification, but quantum noise on Noisy Intermediate-Scale Quantum (NISQ) devices poses practical challenges. This study systematically evaluates how various quantum noise models including dephasing, amplitude damping, depolarizing, thermal noise, bit-flip, and phase-flip affect key QML algorithms (QSVC, Peg-QSVC, QNN, VQC) and feature mapping techniques (ZFeatureMap, ZZFeatureMap, and PauliFeatureMap). Results indicate that QSVC is notably robust under noise, whereas Peg-QSVC and QNN are more sensitive, particularly to depolarizing and amplitude-damping noise. The PauliFeatureMap is especially vulnerable, highlighting difficulties in maintaining accurate classification under noisy conditions. These findings underscore the critical importance of feature map selection and noise mitigation strategies in optimizing QML for genomic classification, with promising implications for personalized medicine.
An Independent Implementation of Quantum Machine Learning Algorithms in Qiskit for Genomic Data
May 16, 2024Abstract:In this paper, we explore the power of Quantum Machine Learning as we extend, implement and evaluate algorithms like Quantum Support Vector Classifier (QSVC), Pegasos-QSVC, Variational Quantum Circuits (VQC), and Quantum Neural Networks (QNN) in Qiskit with diverse feature mapping techniques for genomic sequence classification.
* 2 pager extended abstract
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge