Nate Sauder
Genetic Architect: Discovering Genomic Structure with Learned Neural Architectures
May 23, 2016
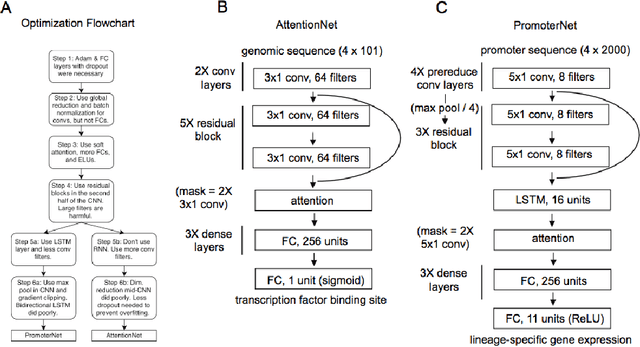
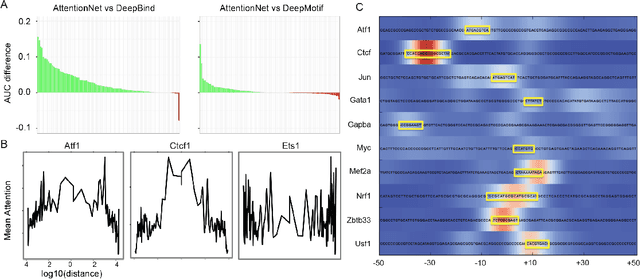
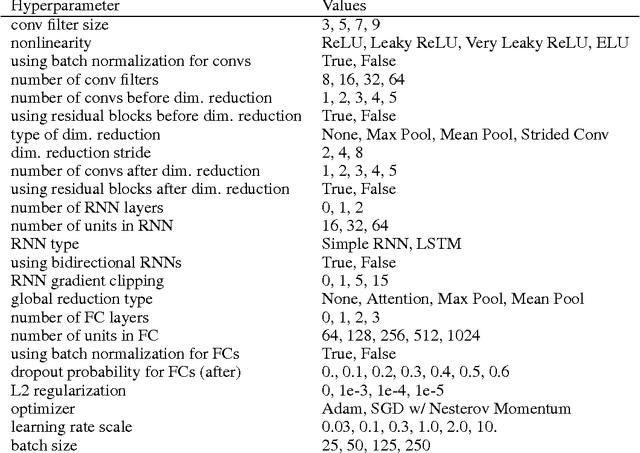
Abstract:Each human genome is a 3 billion base pair set of encoding instructions. Decoding the genome using deep learning fundamentally differs from most tasks, as we do not know the full structure of the data and therefore cannot design architectures to suit it. As such, architectures that fit the structure of genomics should be learned not prescribed. Here, we develop a novel search algorithm, applicable across domains, that discovers an optimal architecture which simultaneously learns general genomic patterns and identifies the most important sequence motifs in predicting functional genomic outcomes. The architectures we find using this algorithm succeed at using only RNA expression data to predict gene regulatory structure, learn human-interpretable visualizations of key sequence motifs, and surpass state-of-the-art results on benchmark genomics challenges.
GradNets: Dynamic Interpolation Between Neural Architectures
Nov 21, 2015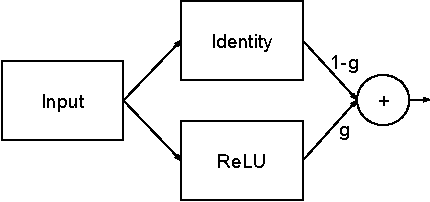
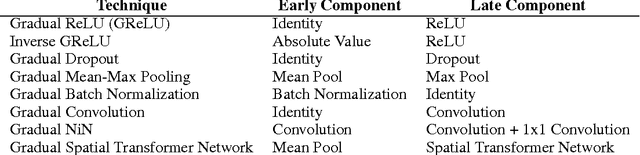

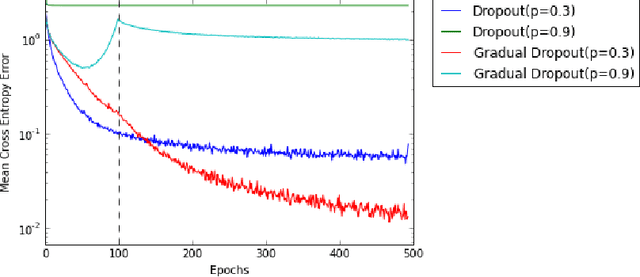
Abstract:In machine learning, there is a fundamental trade-off between ease of optimization and expressive power. Neural Networks, in particular, have enormous expressive power and yet are notoriously challenging to train. The nature of that optimization challenge changes over the course of learning. Traditionally in deep learning, one makes a static trade-off between the needs of early and late optimization. In this paper, we investigate a novel framework, GradNets, for dynamically adapting architectures during training to get the benefits of both. For example, we can gradually transition from linear to non-linear networks, deterministic to stochastic computation, shallow to deep architectures, or even simple downsampling to fully differentiable attention mechanisms. Benefits include increased accuracy, easier convergence with more complex architectures, solutions to test-time execution of batch normalization, and the ability to train networks of up to 200 layers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge