Nadezhda Koriakina
End-to-end Multiple Instance Learning with Gradient Accumulation
Mar 08, 2022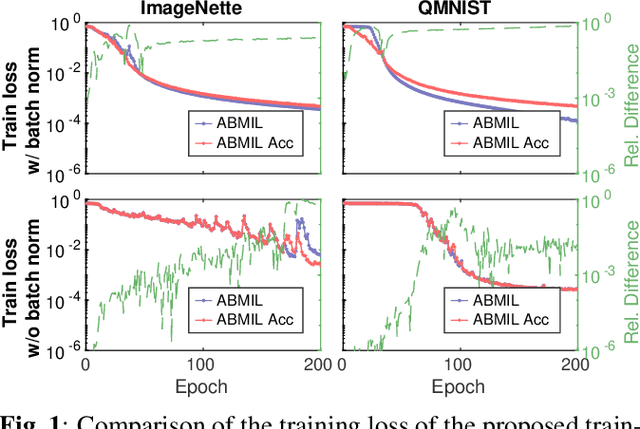
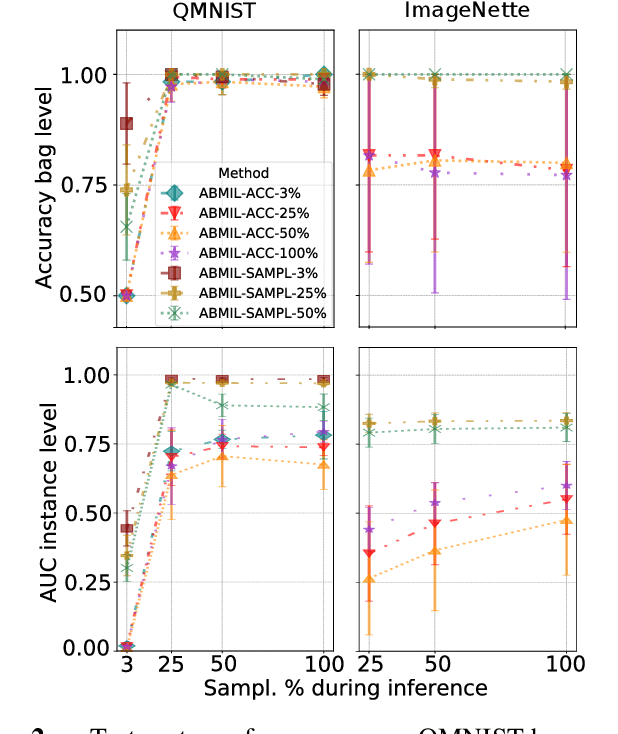
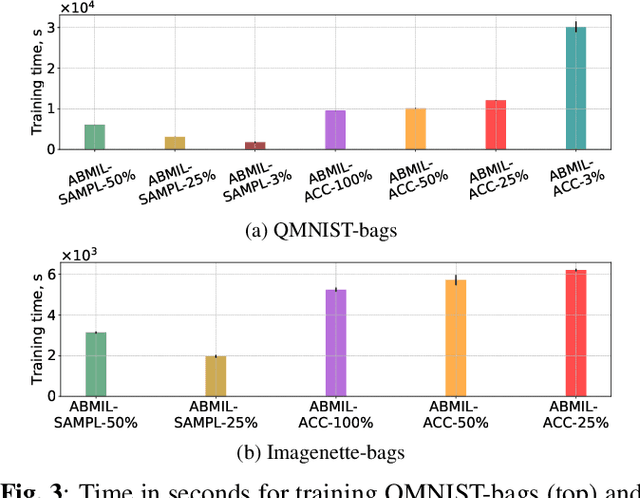
Abstract:Being able to learn on weakly labeled data, and provide interpretability, are two of the main reasons why attention-based deep multiple instance learning (ABMIL) methods have become particularly popular for classification of histopathological images. Such image data usually come in the form of gigapixel-sized whole-slide-images (WSI) that are cropped into smaller patches (instances). However, the sheer size of the data makes training of ABMIL models challenging. All the instances from one WSI cannot be processed at once by conventional GPUs. Existing solutions compromise training by relying on pre-trained models, strategic sampling or selection of instances, or self-supervised learning. We propose a training strategy based on gradient accumulation that enables direct end-to-end training of ABMIL models without being limited by GPU memory. We conduct experiments on both QMNIST and Imagenette to investigate the performance and training time, and compare with the conventional memory-expensive baseline and a recent sampled-based approach. This memory-efficient approach, although slower, reaches performance indistinguishable from the memory-expensive baseline.
Oral cancer detection and interpretation: Deep multiple instance learning versus conventional deep single instance learning
Feb 03, 2022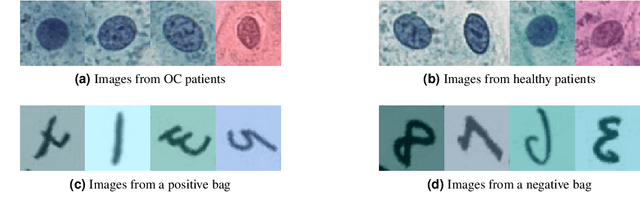
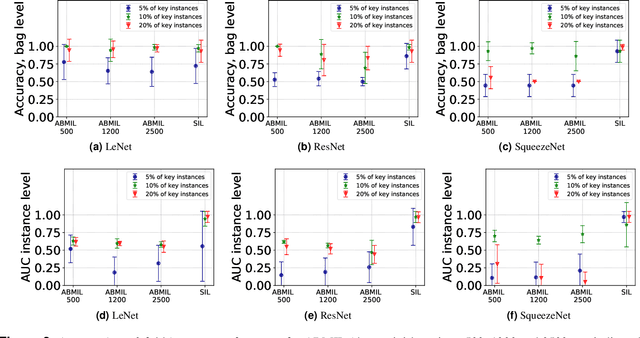
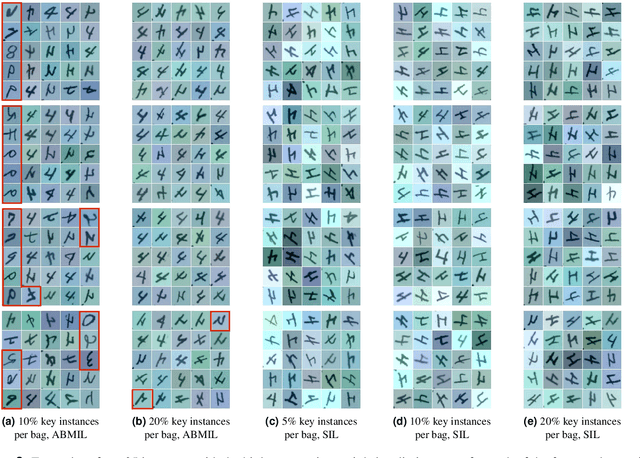
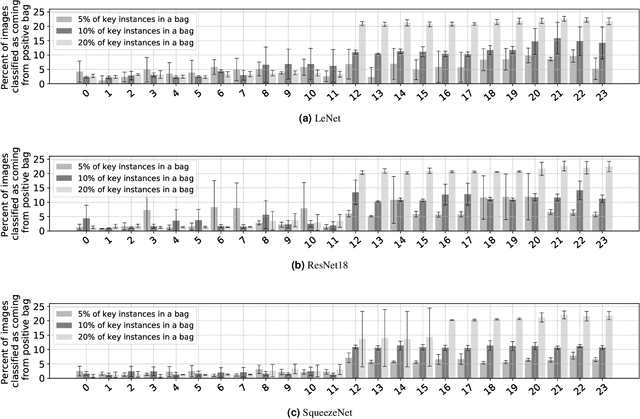
Abstract:The current medical standard for setting an oral cancer (OC) diagnosis is histological examination of a tissue sample from the oral cavity. This process is time consuming and more invasive than an alternative approach of acquiring a brush sample followed by cytological analysis. Skilled cytotechnologists are able to detect changes due to malignancy, however, to introduce this approach into clinical routine is associated with challenges such as a lack of experts and labour-intensive work. To design a trustworthy OC detection system that would assist cytotechnologists, we are interested in AI-based methods that reliably can detect cancer given only per-patient labels (minimizing annotation bias), and also provide information on which cells are most relevant for the diagnosis (enabling supervision and understanding). We, therefore, perform a comparison of a conventional single instance learning (SIL) approach and a modern multiple instance learning (MIL) method suitable for OC detection and interpretation, utilizing three different neural network architectures. To facilitate systematic evaluation of the considered approaches, we introduce a synthetic PAP-QMNIST dataset, that serves as a model of OC data, while offering access to per-instance ground truth. Our study indicates that on PAP-QMNIST, the SIL performs better, on average, than the MIL approach. Performance at the bag level on real-world cytological data is similar for both methods, yet the single instance approach performs better on average. Visual examination by cytotechnologist indicates that the methods manage to identify cells which deviate from normality, including malignant cells as well as those suspicious for dysplasia. We share the code as open source at https://github.com/MIDA-group/OralCancerMILvsSIL
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge