Mikko Impiö
AquaMonitor: A multimodal multi-view image sequence dataset for real-life aquatic invertebrate biodiversity monitoring
May 28, 2025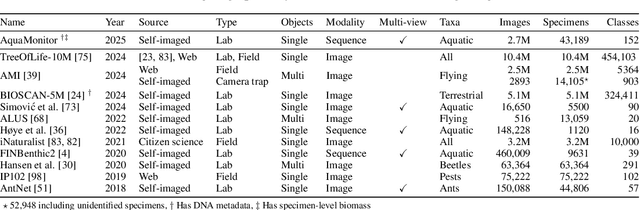
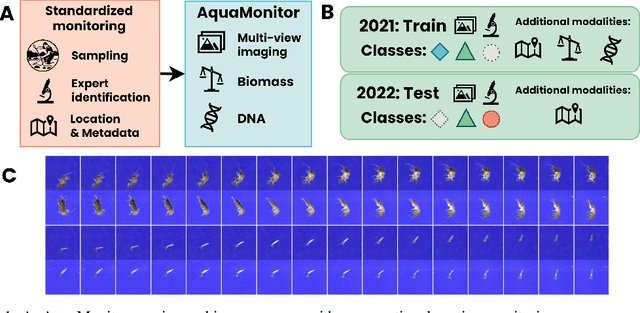
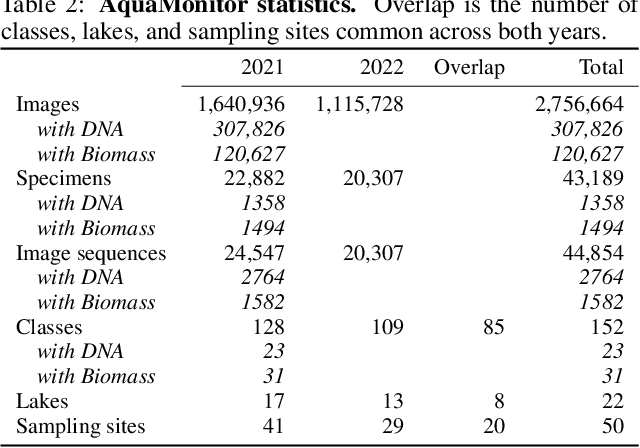
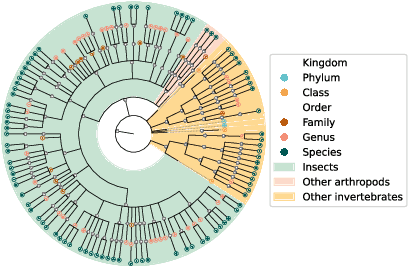
Abstract:This paper presents the AquaMonitor dataset, the first large computer vision dataset of aquatic invertebrates collected during routine environmental monitoring. While several large species identification datasets exist, they are rarely collected using standardized collection protocols, and none focus on aquatic invertebrates, which are particularly laborious to collect. For AquaMonitor, we imaged all specimens from two years of monitoring whenever imaging was possible given practical limitations. The dataset enables the evaluation of automated identification methods for real-life monitoring purposes using a realistically challenging and unbiased setup. The dataset has 2.7M images from 43,189 specimens, DNA sequences for 1358 specimens, and dry mass and size measurements for 1494 specimens, making it also one of the largest biological multi-view and multimodal datasets to date. We define three benchmark tasks and provide strong baselines for these: 1) Monitoring benchmark, reflecting real-life deployment challenges such as open-set recognition, distribution shift, and extreme class imbalance, 2) Classification benchmark, which follows a standard fine-grained visual categorization setup, and 3) Few-shot benchmark, which targets classes with only few training examples from very fine-grained categories. Advancements on the Monitoring benchmark can directly translate to improvement of aquatic biodiversity monitoring, which is an important component of regular legislative water quality assessment in many countries.
Efficient Curation of Invertebrate Image Datasets Using Feature Embeddings and Automatic Size Comparison
Dec 20, 2024Abstract:The amount of image datasets collected for environmental monitoring purposes has increased in the past years as computer vision assisted methods have gained interest. Computer vision applications rely on high-quality datasets, making data curation important. However, data curation is often done ad-hoc and the methods used are rarely published. We present a method for curating large-scale image datasets of invertebrates that contain multiple images of the same taxa and/or specimens and have relatively uniform background in the images. Our approach is based on extracting feature embeddings with pretrained deep neural networks, and using these embeddings to find visually most distinct images by comparing their embeddings to the group prototype embedding. Also, we show that a simple area-based size comparison approach is able to find a lot of common erroneous images, such as images containing detached body parts and misclassified samples. In addition to the method, we propose using novel metrics for evaluating human-in-the-loop outlier detection methods. The implementations of the proposed curation methods, as well as a benchmark dataset containing annotated erroneous images, are publicly available in https://github.com/mikkoim/taxonomist-studio.
Improving Taxonomic Image-based Out-of-distribution Detection With DNA Barcodes
Jun 27, 2024Abstract:Image-based species identification could help scaling biodiversity monitoring to a global scale. Many challenges still need to be solved in order to implement these systems in real-world applications. A reliable image-based monitoring system must detect out-of-distribution (OOD) classes it has not been presented before. This is challenging especially with fine-grained classes. Emerging environmental monitoring techniques, DNA metabarcoding and eDNA, can help by providing information on OOD classes that are present in a sample. In this paper, we study if DNA barcodes can also support in finding the outlier images based on the outlier DNA sequence's similarity to the seen classes. We propose a re-ordering approach that can be easily applied on any pre-trained models and existing OOD detection methods. We experimentally show that the proposed approach improves taxonomic OOD detection compared to all common baselines. We also show that the method works thanks to a correlation between visual similarity and DNA barcode proximity. The code and data are available at https://github.com/mikkoim/dnaimg-ood.
Habitat classification from satellite observations with sparse annotations
Sep 26, 2022

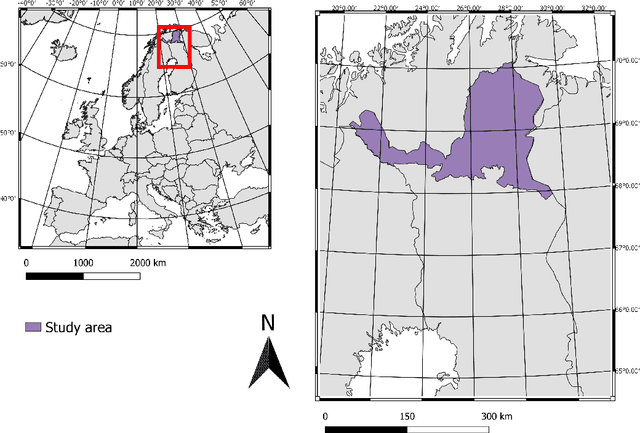

Abstract:Remote sensing benefits habitat conservation by making monitoring of large areas easier compared to field surveying especially if the remote sensed data can be automatically analyzed. An important aspect of monitoring is classifying and mapping habitat types present in the monitored area. Automatic classification is a difficult task, as classes have fine-grained differences and their distributions are long-tailed and unbalanced. Usually training data used for automatic land cover classification relies on fully annotated segmentation maps, annotated from remote sensed imagery to a fairly high-level taxonomy, i.e., classes such as forest, farmland, or urban area. A challenge with automatic habitat classification is that reliable data annotation requires field-surveys. Therefore, full segmentation maps are expensive to produce, and training data is often sparse, point-like, and limited to areas accessible by foot. Methods for utilizing these limited data more efficiently are needed. We address these problems by proposing a method for habitat classification and mapping, and apply this method to classify the entire northern Finnish Lapland area into Natura2000 classes. The method is characterized by using finely-grained, sparse, single-pixel annotations collected from the field, combined with large amounts of unannotated data to produce segmentation maps. Supervised, unsupervised and semi-supervised methods are compared, and the benefits of transfer learning from a larger out-of-domain dataset are demonstrated. We propose a \ac{CNN} biased towards center pixel classification ensembled with a random forest classifier, that produces higher quality classifications than the models themselves alone. We show that cropping augmentations, test-time augmentation and semi-supervised learning can help classification even further.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge