Mehmet Tan
Using Clinical Drug Representations for Improving Mortality and Length of Stay Predictions
Oct 17, 2021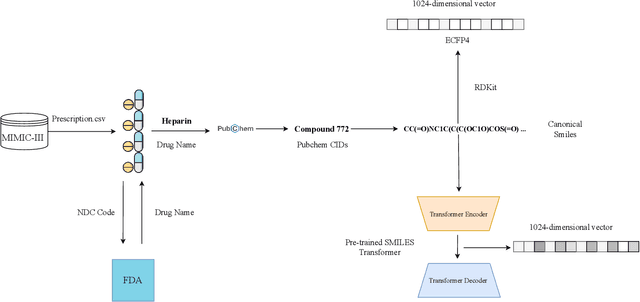
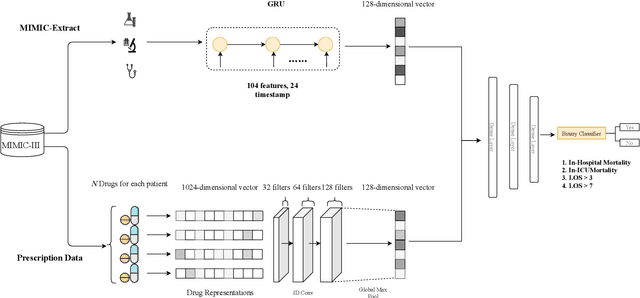
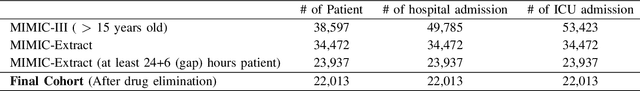
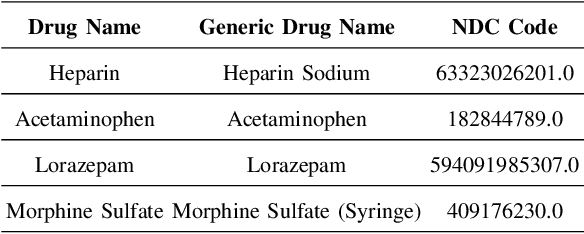
Abstract:Drug representations have played an important role in cheminformatics. However, in the healthcare domain, drug representations have been underused relative to the rest of Electronic Health Record (EHR) data, due to the complexity of high dimensional drug representations and the lack of proper pipeline that will allow to convert clinical drugs to their representations. Time-varying vital signs, laboratory measurements, and related time-series signals are commonly used to predict clinical outcomes. In this work, we demonstrated that using clinical drug representations in addition to other clinical features has significant potential to increase the performance of mortality and length of stay (LOS) models. We evaluate the two different drug representation methods (Extended-Connectivity Fingerprint-ECFP and SMILES-Transformer embedding) on clinical outcome predictions. The results have shown that the proposed multimodal approach achieves substantial enhancement on clinical tasks over baseline models. Using clinical drug representations as additional features improve the LOS prediction for Area Under the Receiver Operating Characteristics (AUROC) around %6 and for Area Under Precision-Recall Curve (AUPRC) by around %5. Furthermore, for the mortality prediction task, there is an improvement of around %2 over the time series baseline in terms of AUROC and %3.5 in terms of AUPRC. The code for the proposed method is available at https://github.com/tanlab/MIMIC-III-Clinical-Drug-Representations.
Improving Clinical Outcome Predictions Using Convolution over Medical Entities with Multimodal Learning
Nov 26, 2020



Abstract:Early prediction of mortality and length of stay(LOS) of a patient is vital for saving a patient's life and management of hospital resources. Availability of electronic health records(EHR) makes a huge impact on the healthcare domain and there has seen several works on predicting clinical problems. However, many studies did not benefit from the clinical notes because of the sparse, and high dimensional nature. In this work, we extract medical entities from clinical notes and use them as additional features besides time-series features to improve our predictions. We propose a convolution based multimodal architecture, which not only learns effectively combining medical entities and time-series ICU signals of patients, but also allows us to compare the effect of different embedding techniques such as Word2vec, FastText on medical entities. In the experiments, our proposed method robustly outperforms all other baseline models including different multimodal architectures for all clinical tasks. The code for the proposed method is available at https://github.com/tanlab/ConvolutionMedicalNer.
Prediction of Drug Synergy by Ensemble Learning
Jan 07, 2020



Abstract:One of the promising methods for the treatment of complex diseases such as cancer is combinational therapy. Due to the combinatorial complexity, machine learning models can be useful in this field, where significant improvements have recently been achieved in determination of synergistic combinations. In this study, we investigate the effectiveness of different compound representations in predicting the drug synergy. On a large drug combination screen dataset, we first demonstrate the use of a promising representation that has not been used for this problem before, then we propose an ensemble on representation-model combinations that outperform each of the baseline models.
Drug response prediction by ensemble learning and drug-induced gene expression signatures
Jul 16, 2018



Abstract:Chemotherapeutic response of cancer cells to a given compound is one of the most fundamental information one requires to design anti-cancer drugs. Recent advances in producing large drug screens against cancer cell lines provided an opportunity to apply machine learning methods for this purpose. In addition to cytotoxicity databases, considerable amount of drug-induced gene expression data has also become publicly available. Following this, several methods that exploit omics data were proposed to predict drug activity on cancer cells. However, due to the complexity of cancer drug mechanisms, none of the existing methods are perfect. One possible direction, therefore, is to combine the strengths of both the methods and the databases for improved performance. We demonstrate that integrating a large number of predictions by the proposed method improves the performance for this task. The predictors in the ensemble differ in several aspects such as the method itself, the number of tasks method considers (multi-task vs. single-task) and the subset of data considered (sub-sampling). We show that all these different aspects contribute to the success of the final ensemble. In addition, we attempt to use the drug screen data together with two novel signatures produced from the drug-induced gene expression profiles of cancer cell lines. Finally, we evaluate the method predictions by in vitro experiments in addition to the tests on data sets.The predictions of the methods, the signatures and the software are available from \url{http://mtan.etu.edu.tr/drug-response-prediction/}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge