Matteo Polsinelli
Multiple Sclerosis Lesions Identification/Segmentation in Magnetic Resonance Imaging using Ensemble CNN and Uncertainty Classification
Aug 26, 2021
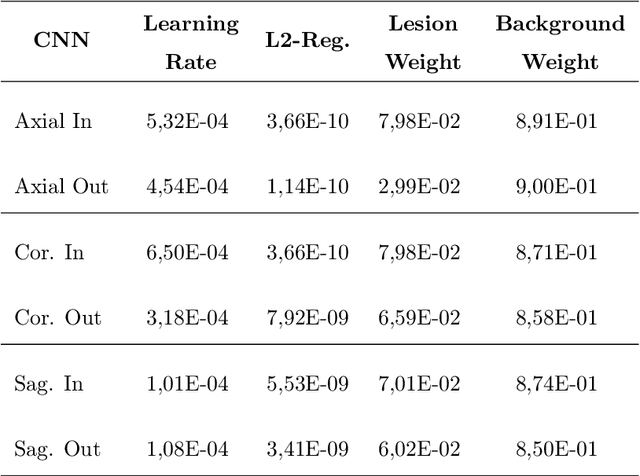
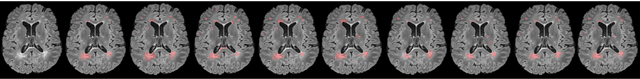

Abstract:To date, several automated strategies for identification/segmentation of Multiple Sclerosis (MS) lesions by Magnetic Resonance Imaging (MRI) have been presented which are either outperformed by human experts or, at least, whose results are well distinguishable from humans. This is due to the ambiguity originated by MRI instabilities, peculiar MS Heterogeneity and MRI unspecific nature with respect to MS. Physicians partially treat the uncertainty generated by ambiguity relying on personal radiological/clinical/anatomical background and experience. We present an automated framework for MS lesions identification/segmentation based on three pivotal concepts to better emulate human reasoning: the modeling of uncertainty; the proposal of two, separately trained, CNN, one optimized with respect to lesions themselves and the other to the environment surrounding lesions, respectively repeated for axial, coronal and sagittal directions; the ensemble of the CNN output. The proposed framework is trained, validated and tested on the 2016 MSSEG benchmark public data set from a single imaging modality, FLuid-Attenuated Inversion Recovery (FLAIR). The comparison, performed on the segmented lesions by means of most of the metrics normally used with respect to the ground-truth and the 7 human raters in MSSEG, prove that there is no significant difference between the proposed framework and the other raters. Results are also shown for the uncertainty, though a comparison with the other raters is impossible.
Convolutional Neural Networks for Automatic Detection of Artifacts from Independent Components Represented in Scalp Topographies of EEG Signals
Sep 08, 2020

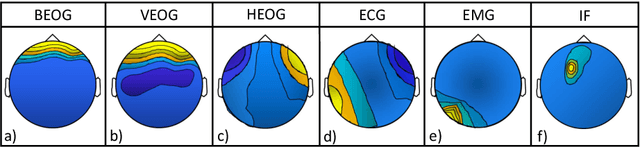
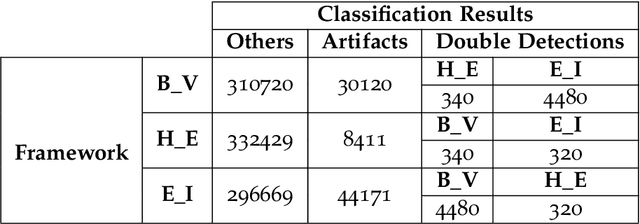
Abstract:Electroencephalography (EEG) measures the electrical brain activity in real-time by using sensors placed on the scalp. Artifacts, due to eye movements and blink, muscular/cardiac activity and generic electrical disturbances, have to be recognized and eliminated to allow a correct interpretation of the useful brain signals (UBS) of EEG. Independent Component Analysis (ICA) is effective to split the signal into independent components (ICs) whose re-projections on 2D scalp topographies (images), also called topoplots, allow to recognize/separate artifacts and by UBS. Until now, IC topoplot analysis, a gold standard in EEG, has been carried on visually by human experts and, hence, not usable in automatic, fast-response EEG. We present a completely automatic and effective framework for EEG artifact recognition by IC topoplots, based on 2D Convolutional Neural Networks (CNNs), capable to divide topoplots in 4 classes: 3 types of artifacts and UBS. The framework setup is described and results are presented, discussed and compared with those obtained by other competitive strategies. Experiments, carried on public EEG datasets, have shown an overall accuracy of above 98%, employing 1.4 sec on a standard PC to classify 32 topoplots, that is to drive an EEG system of 32 sensors. Though not real-time, the proposed framework is efficient enough to be used in fast-response EEG-based Brain-Computer Interfaces (BCI) and faster than other automatic methods based on ICs.
A Light CNN for detecting COVID-19 from CT scans of the chest
Apr 24, 2020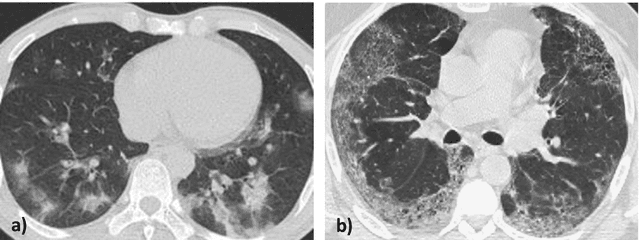
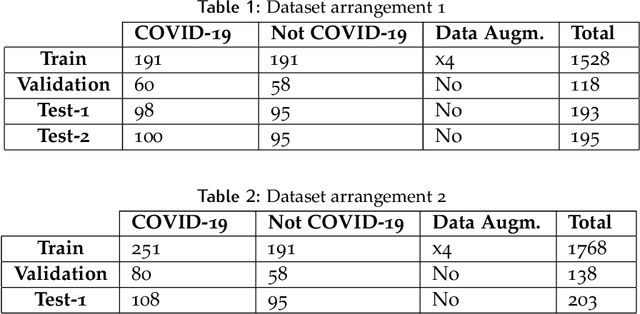
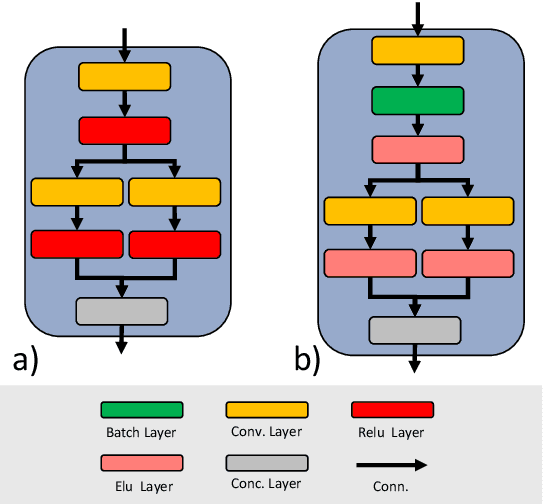
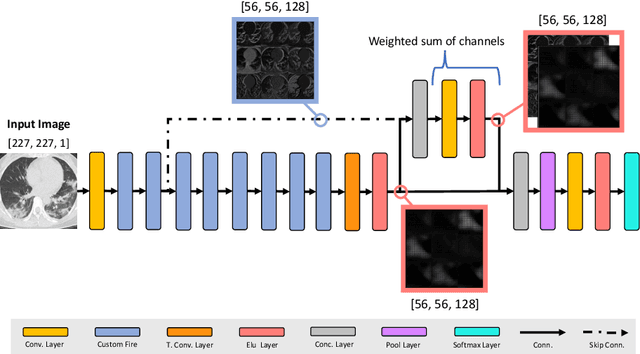
Abstract:OVID-19 is a world-wide disease that has been declared as a pandemic by the World Health Organization. Computer Tomography (CT) imaging of the chest seems to be a valid diagnosis tool to detect COVID-19 promptly and to control the spread of the disease. Deep Learning has been extensively used in medical imaging and convolutional neural networks (CNNs) have been also used for classification of CT images. We propose a light CNN design based on the model of the SqueezeNet, for the efficient discrimination of COVID-19 CT images with other CT images (community-acquired pneumonia and/or healthy images). On the tested datasets, the proposed modified SqueezeNet CNN achieved 83.00\% of accuracy, 85.00\% of sensitivity, 81.00\% of specificity, 81.73\% of precision and 0.8333 of F1Score in a very efficient way (7.81 seconds medium-end laptot without GPU acceleration). Besides performance, the average classification time is very competitive with respect to more complex CNN designs, thus allowing its usability also on medium power computers. In the next future we aim at improving the performances of the method along two directions: 1) by increasing the training dataset (as soon as other CT images will be available); 2) by introducing an efficient pre-processing strategy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge