Mark L. Trew
Basis Function Based Data Driven Learning for the Inverse Problem of Electrocardiography
Feb 01, 2021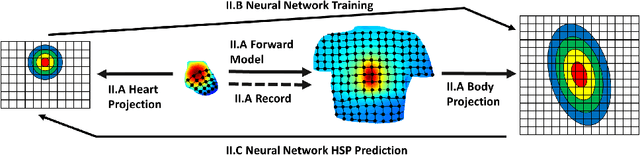
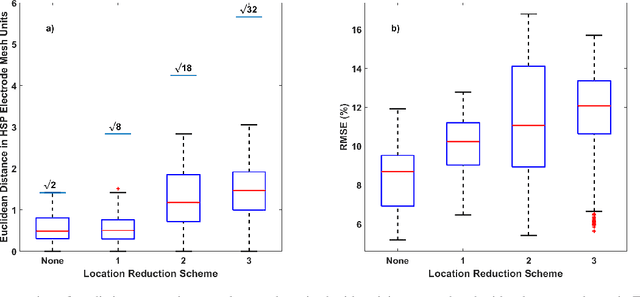
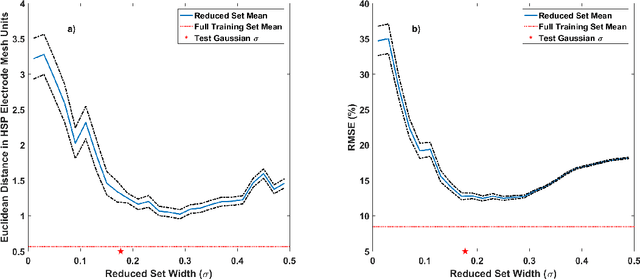
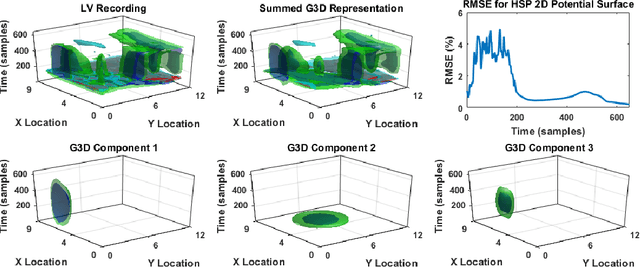
Abstract:Objective: This paper proposes an neural network approach for predicting heart surface potentials (HSPs) from body surface potentials (BSPs), which reframes the traditional inverse problem of electrocardiography into a regression problem through the use of Gaussian 3D (G3D) basis function decomposition. Methods: HSPs were generated using G3D basis functions and passed through a boundary element forward model to obtain corresponding BSPs. The generated BSPs (input) and HSPs (output) were used to train a neural network, which was then used to predict a variety of synthesized and decomposed real-world HSPs. Results: Fitted G3D basis function parameters can accurately reconstruct the real-world left ventricular paced recording with percent root mean squared error (RMSE) of $1.34 \pm 1.30$%. The basis data trained neural network was able to predict G3D basis function synthesized data with RMSE of $8.46 \pm 1.55$%, and G3D representation of real-world data with RMSE of $18.5 \pm 5.25$%. Activation map produced from the predicted time series had a RMSE of 17.0% and mean absolute difference of $10.3 \pm 10.8$ms when compared to that produced from the actual left ventricular paced recording. Conclusion: A Gaussian basis function based data driven model for re-framing the inverse problem of electrocardiography as a regression problem is successful and produces promising time series and activation map predictions of real-world recordings even when only trained using Guassian data. Significance: The HSPs predicted by the neural network can be used to create activation maps to identify cardiac dysfunctions during clinical assessment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge