Marco Verani
Learning cardiac activation and repolarization times with operator learning
May 13, 2025Abstract:Solving partial or ordinary differential equation models in cardiac electrophysiology is a computationally demanding task, particularly when high-resolution meshes are required to capture the complex dynamics of the heart. Moreover, in clinical applications, it is essential to employ computational tools that provide only relevant information, ensuring clarity and ease of interpretation. In this work, we exploit two recently proposed operator learning approaches, namely Fourier Neural Operators (FNO) and Kernel Operator Learning (KOL), to learn the operator mapping the applied stimulus in the physical domain into the activation and repolarization time distributions. These data-driven methods are evaluated on synthetic 2D and 3D domains, as well as on a physiologically realistic left ventricle geometry. Notably, while the learned map between the applied current and activation time has its modelling counterpart in the Eikonal model, no equivalent partial differential equation (PDE) model is known for the map between the applied current and repolarization time. Our results demonstrate that both FNO and KOL approaches are robust to hyperparameter choices and computationally efficient compared to traditional PDE-based Monodomain models. These findings highlight the potential use of these surrogate operators to accelerate cardiac simulations and facilitate their clinical integration.
A model learning framework for inferring the dynamics of transmission rate depending on exogenous variables for epidemic forecasts
Oct 15, 2024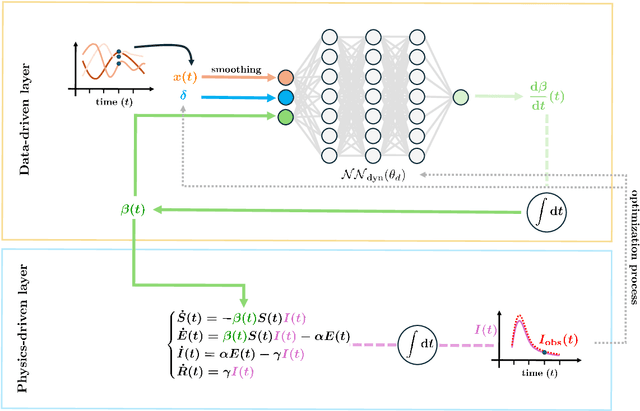
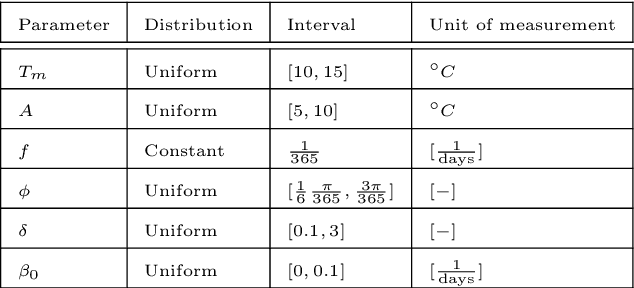
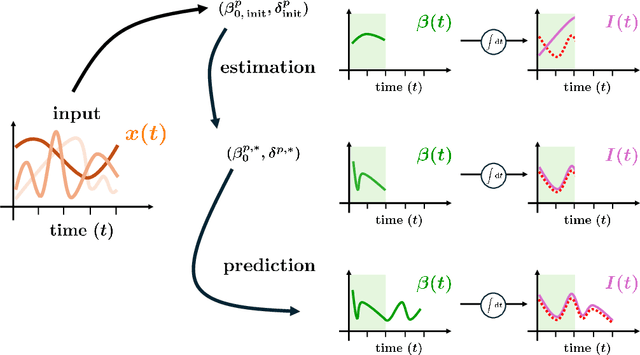
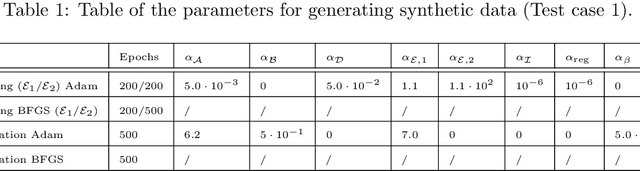
Abstract:In this work, we aim to formalize a novel scientific machine learning framework to reconstruct the hidden dynamics of the transmission rate, whose inaccurate extrapolation can significantly impair the quality of the epidemic forecasts, by incorporating the influence of exogenous variables (such as environmental conditions and strain-specific characteristics). We propose an hybrid model that blends a data-driven layer with a physics-based one. The data-driven layer is based on a neural ordinary differential equation that learns the dynamics of the transmission rate, conditioned on the meteorological data and wave-specific latent parameters. The physics-based layer, instead, consists of a standard SEIR compartmental model, wherein the transmission rate represents an input. The learning strategy follows an end-to-end approach: the loss function quantifies the mismatch between the actual numbers of infections and its numerical prediction obtained from the SEIR model incorporating as an input the transmission rate predicted by the neural ordinary differential equation. We validate this original approach using both a synthetic test case and a realistic test case based on meteorological data (temperature and humidity) and influenza data from Italy between 2010 and 2020. In both scenarios, we achieve low generalization error on the test set and observe strong alignment between the reconstructed model and established findings on the influence of meteorological factors on epidemic spread. Finally, we implement a data assimilation strategy to adapt the neural equation to the specific characteristics of an epidemic wave under investigation, and we conduct sensitivity tests on the network hyperparameters.
Learning epidemic trajectories through Kernel Operator Learning: from modelling to optimal control
Apr 17, 2024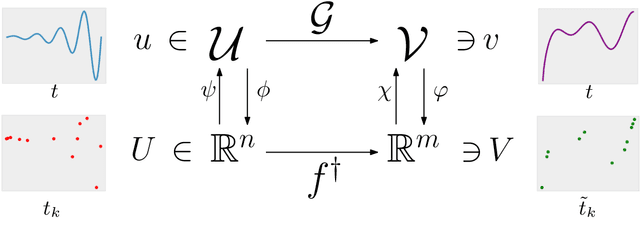
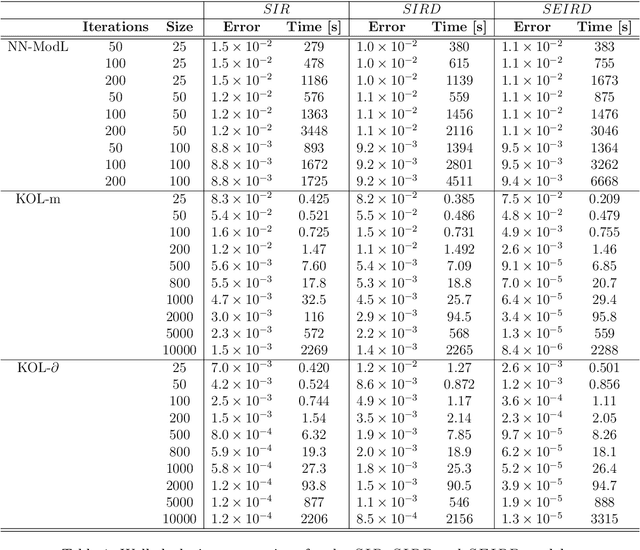
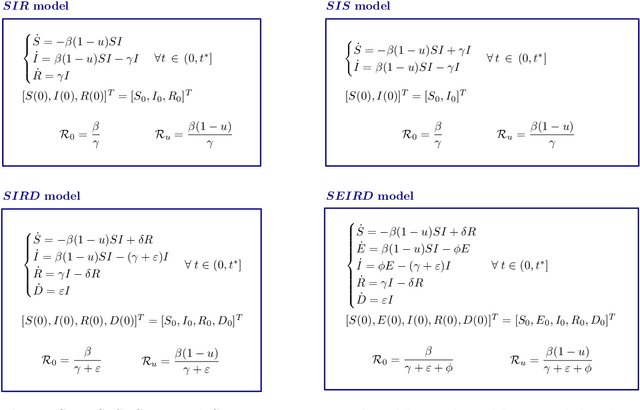
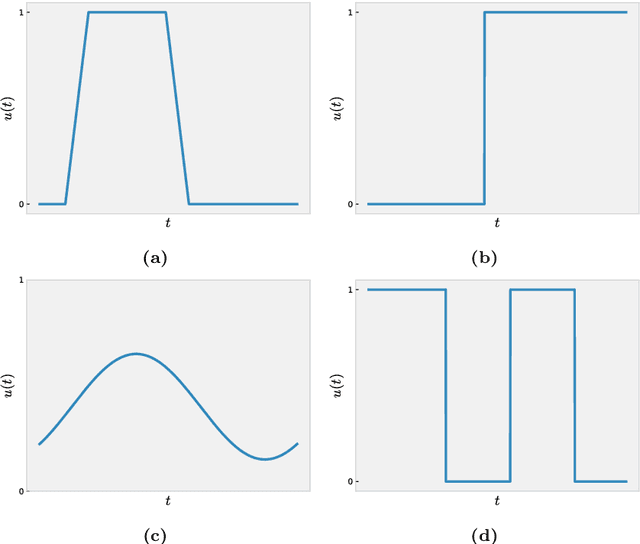
Abstract:Since infectious pathogens start spreading into a susceptible population, mathematical models can provide policy makers with reliable forecasts and scenario analyses, which can be concretely implemented or solely consulted. In these complex epidemiological scenarios, machine learning architectures can play an important role, since they directly reconstruct data-driven models circumventing the specific modelling choices and the parameter calibration, typical of classical compartmental models. In this work, we discuss the efficacy of Kernel Operator Learning (KOL) to reconstruct population dynamics during epidemic outbreaks, where the transmission rate is ruled by an input strategy. In particular, we introduce two surrogate models, named KOL-m and KOL-$\partial$, which reconstruct in two different ways the evolution of the epidemics. Moreover, we evaluate the generalization performances of the two approaches with different kernels, including the Neural Tangent Kernels, and compare them with a classical neural network model learning method. Employing synthetic but semi-realistic data, we show how the two introduced approaches are suitable for realizing fast and robust forecasts and scenario analyses, and how these approaches are competitive for determining optimal intervention strategies with respect to specific performance measures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge