Manoj Raju
Q-Map: Clinical Concept Mining from Clinical Documents
Jul 16, 2018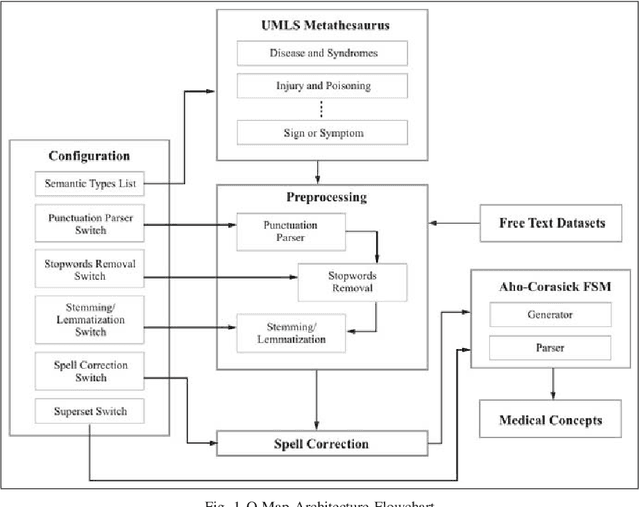
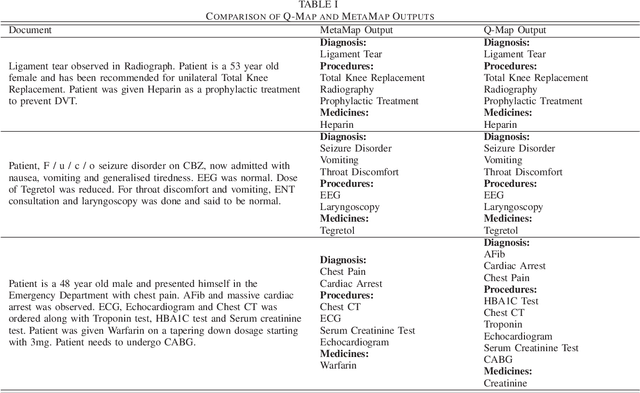
Abstract:Over the past decade, there has been a steep rise in the data-driven analysis in major areas of medicine, such as clinical decision support system, survival analysis, patient similarity analysis, image analytics etc. Most of the data in the field are well-structured and available in numerical or categorical formats which can be used for experiments directly. But on the opposite end of the spectrum, there exists a wide expanse of data that is intractable for direct analysis owing to its unstructured nature which can be found in the form of discharge summaries, clinical notes, procedural notes which are in human written narrative format and neither have any relational model nor any standard grammatical structure. An important step in the utilization of these texts for such studies is to transform and process the data to retrieve structured information from the haystack of irrelevant data using information retrieval and data mining techniques. To address this problem, the authors present Q-Map in this paper, which is a simple yet robust system that can sift through massive datasets with unregulated formats to retrieve structured information aggressively and efficiently. It is backed by an effective mining technique which is based on a string matching algorithm that is indexed on curated knowledge sources, that is both fast and configurable. The authors also briefly examine its comparative performance with MetaMap, one of the most reputed tools for medical concepts retrieval and present the advantages the former displays over the latter.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge