Mahdi Tabassian
Deep Spatiotemporal Clutter Filtering of Transthoracic Echocardiographic Images Using a 3D Convolutional Auto-Encoder
Jan 23, 2024

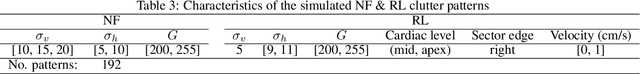

Abstract:This study presents a deep convolutional auto-encoder network for filtering reverberation artifacts, from transthoracic echocardiographic (TTE) image sequences. Given the spatiotemporal nature of these artifacts, the filtering network was built using 3D convolutional layers to suppress the clutter patterns throughout the cardiac cycle. The network was designed by taking advantage of: i) an attention mechanism to focus primarily on cluttered regions and ii) residual learning to preserve fine structures of the image frames. To train the deep network, a diverse set of artifact patterns was simulated and the simulated patterns were superimposed onto artifact-free ultra-realistic synthetic TTE sequences of six ultrasound vendors to generate input of the filtering network. The artifact-free sequences served as ground-truth. Performance of the filtering network was evaluated using unseen synthetic as well as in-vivo artifactual sequences. Satisfactory results obtained using the latter dataset confirmed the good generalization performance of the proposed network which was trained using the synthetic sequences and simulated artifact patterns. Suitability of the clutter-filtered sequences for further processing was assessed by computing segmental strain curves from them. The results showed that the large discrepancy between the strain profiles computed from the cluttered segments and their corresponding segments in the clutter-free images was significantly reduced after filtering the sequences using the proposed network. The trained deep network could process an artifactual TTE sequence in a fraction of a second and can be used for real-time clutter filtering. Moreover, it can improve the precision of the clinical indexes that are computed from the TTE sequences. The source code of the proposed method is available at: https://github.com/MahdiTabassian/Deep-Clutter-Filtering/tree/main.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge