Luca Masera
Multiple protein feature prediction with statistical relational learning
Sep 27, 2016

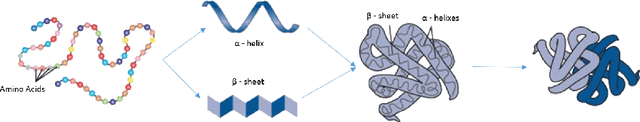
Abstract:High throughput sequencing techniques have highly impactedon modern biology, widening the gap between sequenced andannotated data. Automatic annotation tools are thereforeof the foremost importance to guide biologists' experiments. However, most of the state-of-the-art methods rely on annotation transfer, offering reliable predictions only in homology settings. In this work we present a novel appraoch to protein feature prediction, which exploits the Semanti Based Regularization to inject prior knowledge in the learning process. The experimental results conducted on the yeast genome show that the introduction of the constraints positively impacts on the overall prediction quality.
Very Simple Classifier: a Concept Binary Classifier toInvestigate Features Based on Subsampling and Localility
Sep 14, 2016
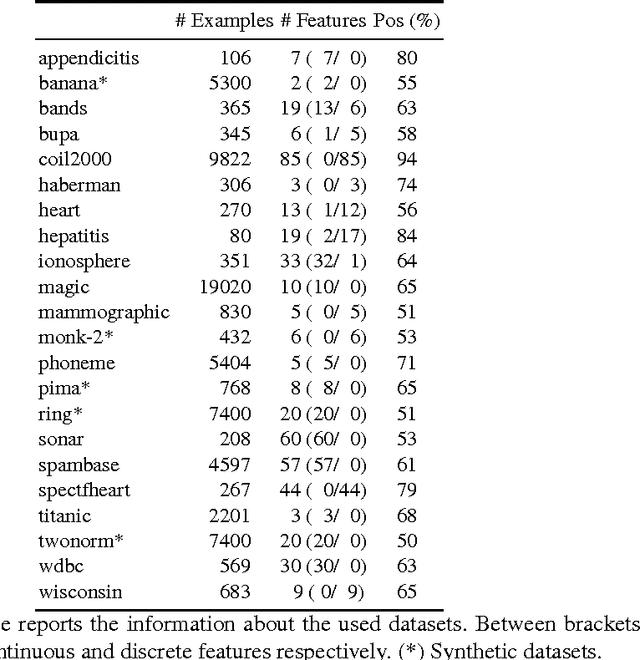
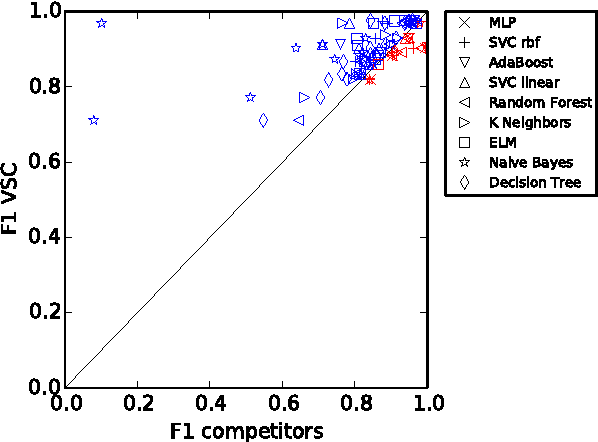
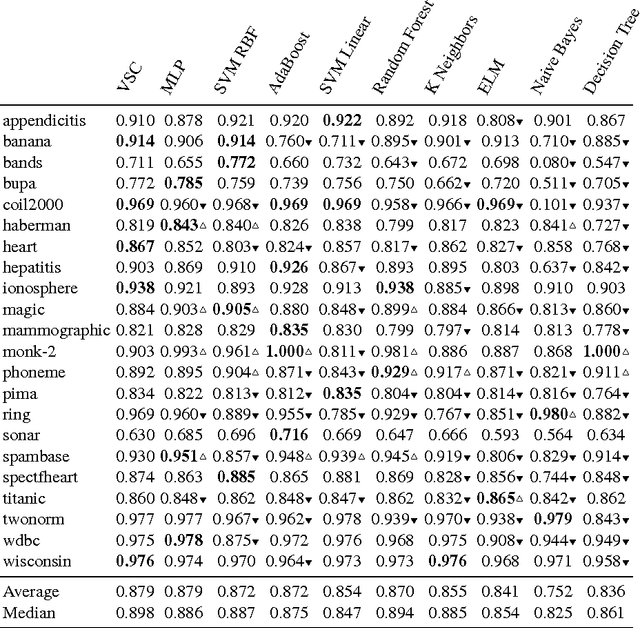
Abstract:We propose Very Simple Classifier (VSC) a novel method designed to incorporate the concepts of subsampling and locality in the definition of features to be used as the input of a perceptron. The rationale is that locality theoretically guarantees a bound on the generalization error. Each feature in VSC is a max-margin classifier built on randomly-selected pairs of samples. The locality in VSC is achieved by multiplying the value of the feature by a confidence measure that can be characterized in terms of the Chebichev inequality. The output of the layer is then fed in a output layer of neurons. The weights of the output layer are then determined by a regularized pseudoinverse. Extensive comparison of VSC against 9 competitors in the task of binary classification is carried out. Results on 22 benchmark datasets with fixed parameters show that VSC is competitive with the Multi Layer Perceptron (MLP) and outperforms the other competitors. An exploration of the parameter space shows VSC can outperform MLP.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge