Linjia Kang
Learning with Challenges: Adaptive Difficulty-Aware Data Generation for Mobile GUI Agent Training
Jan 30, 2026Abstract:Large-scale, high-quality interaction trajectories are essential for advancing mobile Graphical User Interface (GUI) agents. While existing methods typically rely on labor-intensive human demonstrations or automated model exploration to generate GUI trajectories, they lack fine-grained control over task difficulty. This fundamentally restricts learning effectiveness due to the mismatch between the training difficulty and the agent's capabilities. Inspired by how humans acquire skills through progressively challenging tasks, we propose MobileGen, a novel data generation framework that adaptively aligns training difficulty with the GUI agent's capability frontier. Specifically, MobileGen explicitly decouples task difficulty into structural (e.g., trajectory length) and semantic (e.g., task goal) dimensions. It then iteratively evaluates the agent on a curated prior dataset to construct a systematic profile of its capability frontier across these two dimensions. With this profile, the probability distribution of task difficulty is adaptively computed, from which the target difficulty for the next round of training can be sampled. Guided by the sampled difficulty, a multi-agent controllable generator is finally used to synthesize high-quality interaction trajectories along with corresponding task instructions. Extensive experiments show that MobileGen consistently outperforms existing data generation methods by improving the average performance of GUI agents by 1.57 times across multiple challenging benchmarks. This highlights the importance of capability-aligned data generation for effective mobile GUI agent training.
CoBel-World: Harnessing LLM Reasoning to Build a Collaborative Belief World for Optimizing Embodied Multi-Agent Collaboration
Sep 26, 2025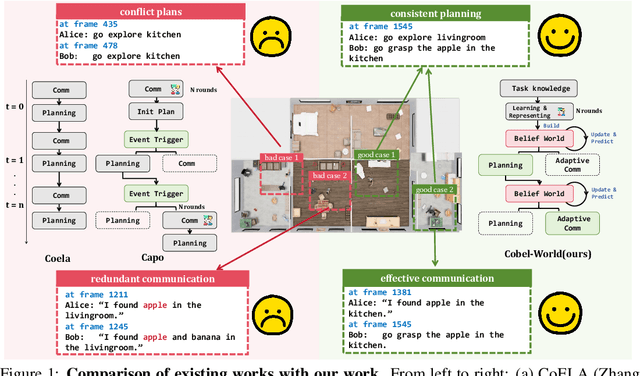
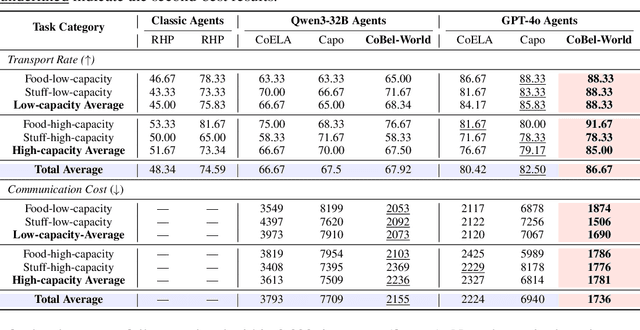
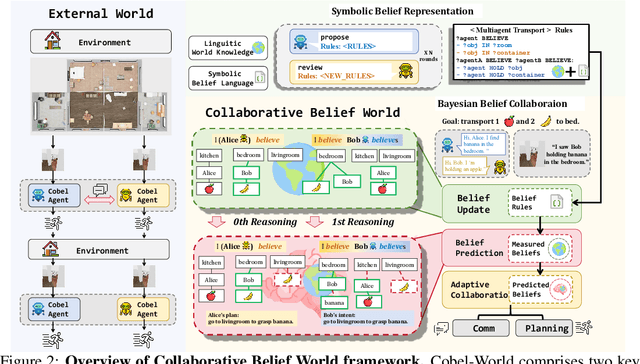
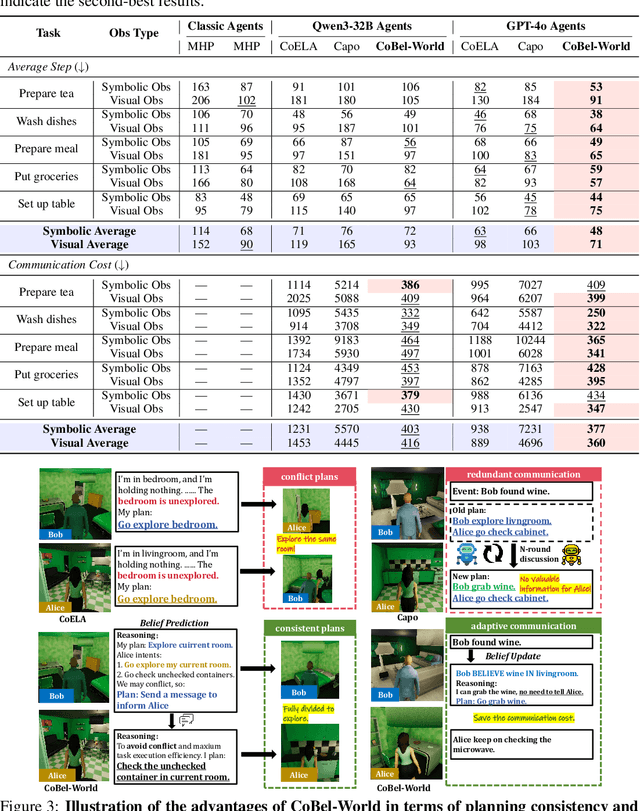
Abstract:Effective real-world multi-agent collaboration requires not only accurate planning but also the ability to reason about collaborators' intents -- a crucial capability for avoiding miscoordination and redundant communication under partial observable environments. Due to their strong planning and reasoning capabilities, large language models (LLMs) have emerged as promising autonomous agents for collaborative task solving. However, existing collaboration frameworks for LLMs overlook their reasoning potential for dynamic intent inference, and thus produce inconsistent plans and redundant communication, reducing collaboration efficiency. To bridge this gap, we propose CoBel-World, a novel framework that equips LLM agents with a collaborative belief world -- an internal representation jointly modeling the physical environment and collaborators' mental states. CoBel-World enables agents to parse open-world task knowledge into structured beliefs via a symbolic belief language, and perform zero-shot Bayesian-style belief updates through LLM reasoning. This allows agents to proactively detect potential miscoordination (e.g., conflicting plans) and communicate adaptively. Evaluated on challenging embodied benchmarks (i.e., TDW-MAT and C-WAH), CoBel-World significantly reduces communication costs by 22-60% and improves task completion efficiency by 4-28% compared to the strongest baseline. Our results show that explicit, intent-aware belief modeling is essential for efficient and human-like collaboration in LLM-based multi-agent systems.
Adapting Differential Molecular Representation with Hierarchical Prompts for Multi-label Property Prediction
May 29, 2024



Abstract:Accurate prediction of molecular properties is critical in the field of drug discovery. However, existing methods do not fully consider the fact that molecules in the real world usually possess multiple property labels, and complex high-order relationships may exist among these labels. Therefore, molecular representation learning models should generate differential molecular representations that consider multi-granularity correlation information among tasks. To this end, our research introduces a Hierarchical Prompted Molecular Representation Learning Framework (HiPM), which enhances the differential expression of tasks in molecular representations through task-aware prompts, and utilizes shared information among labels to mitigate negative transfer between different tasks. HiPM primarily consists of two core components: the Molecular Representation Encoder (MRE) and the Task-Aware Prompter (TAP). The MRE employs a hierarchical message-passing network architecture to capture molecular features at both the atomic and motif levels, while the TAP uses agglomerative hierarchical clustering to build a prompt tree that reflects the affinity and distinctiveness of tasks, enabling the model to effectively handle the complexity of multi-label property predictions. Extensive experiments demonstrate that HiPM achieves state-of-the-art performance across various multi-label datasets, offering a new perspective on multi-label molecular representation learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge