Leontios J. Hadjileontiadis
Variational decomposition autoencoding improves disentanglement of latent representations
Jan 11, 2026Abstract:Understanding the structure of complex, nonstationary, high-dimensional time-evolving signals is a central challenge in scientific data analysis. In many domains, such as speech and biomedical signal processing, the ability to learn disentangled and interpretable representations is critical for uncovering latent generative mechanisms. Traditional approaches to unsupervised representation learning, including variational autoencoders (VAEs), often struggle to capture the temporal and spectral diversity inherent in such data. Here we introduce variational decomposition autoencoding (VDA), a framework that extends VAEs by incorporating a strong structural bias toward signal decomposition. VDA is instantiated through variational decomposition autoencoders (DecVAEs), i.e., encoder-only neural networks that combine a signal decomposition model, a contrastive self-supervised task, and variational prior approximation to learn multiple latent subspaces aligned with time-frequency characteristics. We demonstrate the effectiveness of DecVAEs on simulated data and three publicly available scientific datasets, spanning speech recognition, dysarthria severity evaluation, and emotional speech classification. Our results demonstrate that DecVAEs surpass state-of-the-art VAE-based methods in terms of disentanglement quality, generalization across tasks, and the interpretability of latent encodings. These findings suggest that decomposition-aware architectures can serve as robust tools for extracting structured representations from dynamic signals, with potential applications in clinical diagnostics, human-computer interaction, and adaptive neurotechnologies.
VarCoNet: A variability-aware self-supervised framework for functional connectome extraction from resting-state fMRI
Oct 02, 2025



Abstract:Accounting for inter-individual variability in brain function is key to precision medicine. Here, by considering functional inter-individual variability as meaningful data rather than noise, we introduce VarCoNet, an enhanced self-supervised framework for robust functional connectome (FC) extraction from resting-state fMRI (rs-fMRI) data. VarCoNet employs self-supervised contrastive learning to exploit inherent functional inter-individual variability, serving as a brain function encoder that generates FC embeddings readily applicable to downstream tasks even in the absence of labeled data. Contrastive learning is facilitated by a novel augmentation strategy based on segmenting rs-fMRI signals. At its core, VarCoNet integrates a 1D-CNN-Transformer encoder for advanced time-series processing, enhanced with a robust Bayesian hyperparameter optimization. Our VarCoNet framework is evaluated on two downstream tasks: (i) subject fingerprinting, using rs-fMRI data from the Human Connectome Project, and (ii) autism spectrum disorder (ASD) classification, using rs-fMRI data from the ABIDE I and ABIDE II datasets. Using different brain parcellations, our extensive testing against state-of-the-art methods, including 13 deep learning methods, demonstrates VarCoNet's superiority, robustness, interpretability, and generalizability. Overall, VarCoNet provides a versatile and robust framework for FC analysis in rs-fMRI.
CochCeps-Augment: A Novel Self-Supervised Contrastive Learning Using Cochlear Cepstrum-based Masking for Speech Emotion Recognition
Feb 10, 2024
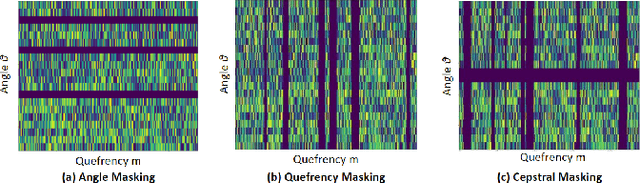
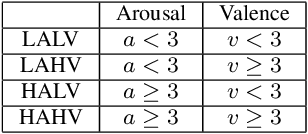
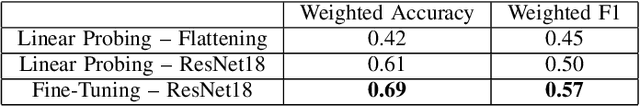
Abstract:Self-supervised learning (SSL) for automated speech recognition in terms of its emotional content, can be heavily degraded by the presence noise, affecting the efficiency of modeling the intricate temporal and spectral informative structures of speech. Recently, SSL on large speech datasets, as well as new audio-specific SSL proxy tasks, such as, temporal and frequency masking, have emerged, yielding superior performance compared to classic approaches drawn from the image augmentation domain. Our proposed contribution builds upon this successful paradigm by introducing CochCeps-Augment, a novel bio-inspired masking augmentation task for self-supervised contrastive learning of speech representations. Specifically, we utilize the newly introduced bio-inspired cochlear cepstrogram (CCGRAM) to derive noise robust representations of input speech, that are then further refined through a self-supervised learning scheme. The latter employs SimCLR to generate contrastive views of a CCGRAM through masking of its angle and quefrency dimensions. Our experimental approach and validations on the emotion recognition K-EmoCon benchmark dataset, for the first time via a speaker-independent approach, features unsupervised pre-training, linear probing and fine-tuning. Our results potentiate CochCeps-Augment to serve as a standard tool in speech emotion recognition analysis, showing the added value of incorporating bio-inspired masking as an informative augmentation task for self-supervision. Our code for implementing CochCeps-Augment will be made available at: https://github.com/GiannisZgs/CochCepsAugment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge