Leila Bagheriye
Advancements in Real-Time Oncology Diagnosis: Harnessing AI and Image Fusion Techniques
Mar 14, 2025Abstract:Real-time computer-aided diagnosis using artificial intelligence (AI), with images, can help oncologists diagnose cancer with high accuracy and in an early phase. We reviewed real-time AI-based analyzed images for decision-making in different cancer types. This paper provides insights into the present and future potential of real-time imaging and image fusion. It explores various real-time techniques, encompassing technical solutions, AI-based imaging, and image fusion diagnosis across multiple anatomical areas, and electromagnetic needle tracking. To provide a thorough overview, this paper discusses ultrasound image fusion, real-time in vivo cancer diagnosis with different spectroscopic techniques, different real-time optical imaging-based cancer diagnosis techniques, elastography-based cancer diagnosis, cervical cancer detection using neuromorphic architectures, different fluorescence image-based cancer diagnosis techniques, and hyperspectral imaging-based cancer diagnosis. We close by offering a more futuristic overview to solve existing problems in real-time image-based cancer diagnosis.
STAL: Spike Threshold Adaptive Learning Encoder for Classification of Pain-Related Biosignal Data
Jul 11, 2024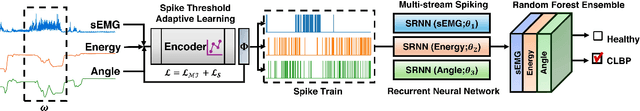
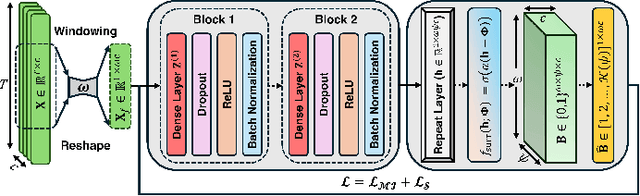
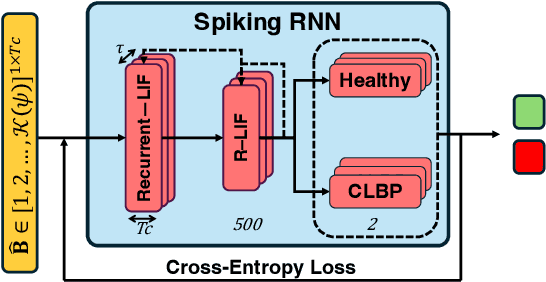
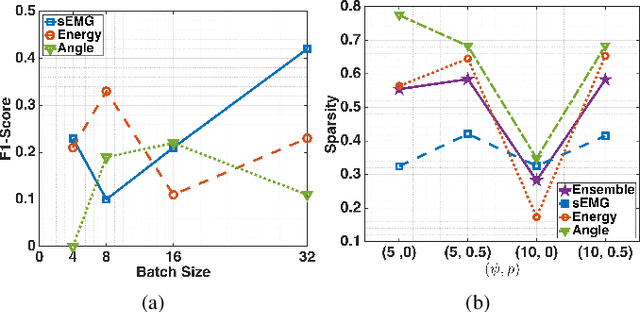
Abstract:This paper presents the first application of spiking neural networks (SNNs) for the classification of chronic lower back pain (CLBP) using the EmoPain dataset. Our work has two main contributions. We introduce Spike Threshold Adaptive Learning (STAL), a trainable encoder that effectively converts continuous biosignals into spike trains. Additionally, we propose an ensemble of Spiking Recurrent Neural Network (SRNN) classifiers for the multi-stream processing of sEMG and IMU data. To tackle the challenges of small sample size and class imbalance, we implement minority over-sampling with weighted sample replacement during batch creation. Our method achieves outstanding performance with an accuracy of 80.43%, AUC of 67.90%, F1 score of 52.60%, and Matthews Correlation Coefficient (MCC) of 0.437, surpassing traditional rate-based and latency-based encoding methods. The STAL encoder shows superior performance in preserving temporal dynamics and adapting to signal characteristics. Importantly, our approach (STAL-SRNN) outperforms the best deep learning method in terms of MCC, indicating better balanced class prediction. This research contributes to the development of neuromorphic computing for biosignal analysis. It holds promise for energy-efficient, wearable solutions in chronic pain management.
Cancer Subtype Identification through Integrating Inter and Intra Dataset Relationships in Multi-Omics Data
Dec 02, 2023Abstract:The integration of multi-omics data has emerged as a promising approach for gaining comprehensive insights into complex diseases such as cancer. This paper proposes a novel approach to identify cancer subtypes through the integration of multi-omics data for clustering. The proposed method, named LIDAF utilises affinity matrices based on linear relationships between and within different omics datasets (Linear Inter and Intra Dataset Affinity Fusion (LIDAF)). Canonical Correlation Analysis is in this paper employed to create distance matrices based on Euclidean distances between canonical variates. The distance matrices are converted to affinity matrices and those are fused in a three-step process. The proposed LIDAF addresses the limitations of the existing method resulting in improvement of clustering performance as measured by the Adjusted Rand Index and the Normalized Mutual Information score. Moreover, our proposed LIDAF approach demonstrates a notable enhancement in 50% of the log10 rank p-values obtained from Cox survival analysis, surpassing the performance of the best reported method, highlighting its potential of identifying distinct cancer subtypes.
Bayesian Integration of Information Using Top-Down Modulated WTA Networks
Aug 29, 2023Abstract:Winner Take All (WTA) circuits a type of Spiking Neural Networks (SNN) have been suggested as facilitating the brain's ability to process information in a Bayesian manner. Research has shown that WTA circuits are capable of approximating hierarchical Bayesian models via Expectation Maximization (EM). So far, research in this direction has focused on bottom up processes. This is contrary to neuroscientific evidence that shows that, besides bottom up processes, top down processes too play a key role in information processing by the human brain. Several functions ascribed to top down processes include direction of attention, adjusting for expectations, facilitation of encoding and recall of learned information, and imagery. This paper explores whether WTA circuits are suitable for further integrating information represented in separate WTA networks. Furthermore, it explores whether, and under what circumstances, top down processes can improve WTA network performance with respect to inference and learning. The results show that WTA circuits are capable of integrating the probabilistic information represented by other WTA networks, and that top down processes can improve a WTA network's inference and learning performance. Notably, it is able to do this according to key neuromorphic principles, making it ideal for low-latency and energy efficient implementation on neuromorphic hardware.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge