Leevi Annala
Adaptive Variance Thresholding: A Novel Approach to Improve Existing Deep Transfer Vision Models and Advance Automatic Knee-Joint Osteoarthritis Classification
Nov 10, 2023



Abstract:Knee-Joint Osteoarthritis (KOA) is a prevalent cause of global disability and is inherently complex to diagnose due to its subtle radiographic markers and individualized progression. One promising classification avenue involves applying deep learning methods; however, these techniques demand extensive, diversified datasets, which pose substantial challenges due to medical data collection restrictions. Existing practices typically resort to smaller datasets and transfer learning. However, this approach often inherits unnecessary pre-learned features that can clutter the classifier's vector space, potentially hampering performance. This study proposes a novel paradigm for improving post-training specialized classifiers by introducing adaptive variance thresholding (AVT) followed by Neural Architecture Search (NAS). This approach led to two key outcomes: an increase in the initial accuracy of the pre-trained KOA models and a 60-fold reduction in the NAS input vector space, thus facilitating faster inference speed and a more efficient hyperparameter search. We also applied this approach to an external model trained for KOA classification. Despite its initial performance, the application of our methodology improved its average accuracy, making it one of the top three KOA classification models.
Synthesizing Bidirectional Temporal States of Knee Osteoarthritis Radiographs with Cycle-Consistent Generative Adversarial Neural Networks
Nov 10, 2023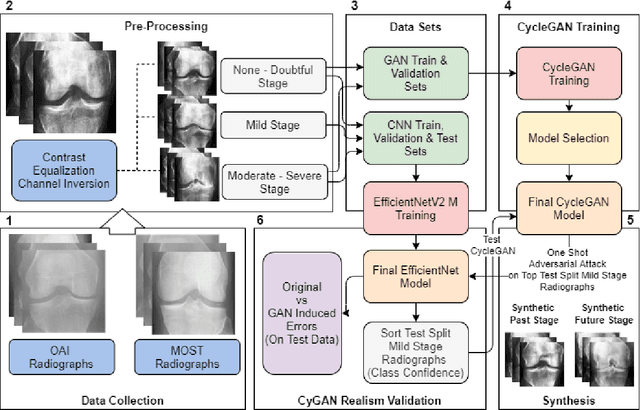
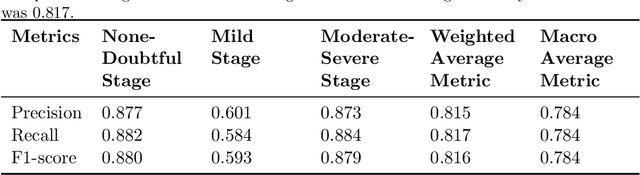
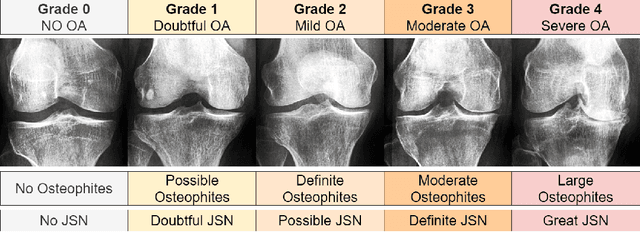
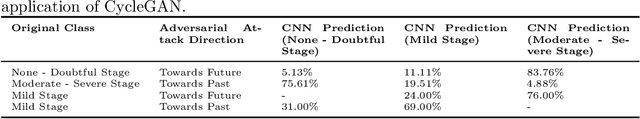
Abstract:Knee Osteoarthritis (KOA), a leading cause of disability worldwide, is challenging to detect early due to subtle radiographic indicators. Diverse, extensive datasets are needed but are challenging to compile because of privacy, data collection limitations, and the progressive nature of KOA. However, a model capable of projecting genuine radiographs into different OA stages could augment data pools, enhance algorithm training, and offer pre-emptive prognostic insights. In this study, we trained a CycleGAN model to synthesize past and future stages of KOA on any genuine radiograph. The model was validated using a Convolutional Neural Network that was deceived into misclassifying disease stages in transformed images, demonstrating the CycleGAN's ability to effectively transform disease characteristics forward or backward in time. The model was particularly effective in synthesizing future disease states and showed an exceptional ability to retroactively transition late-stage radiographs to earlier stages by eliminating osteophytes and expanding knee joint space, signature characteristics of None or Doubtful KOA. The model's results signify a promising potential for enhancing diagnostic models, data augmentation, and educational and prognostic usage in healthcare. Nevertheless, further refinement, validation, and a broader evaluation process encompassing both CNN-based assessments and expert medical feedback are emphasized for future research and development.
Exploring the Efficacy of Base Data Augmentation Methods in Deep Learning-Based Radiograph Classification of Knee Joint Osteoarthritis
Nov 10, 2023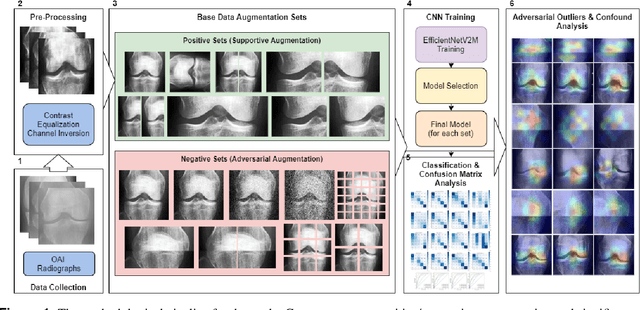

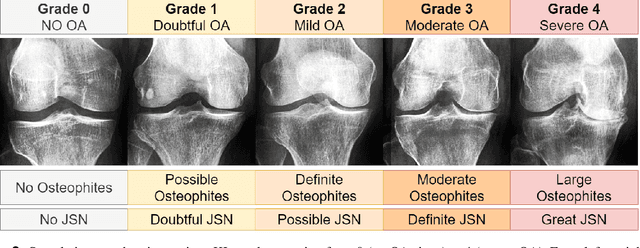
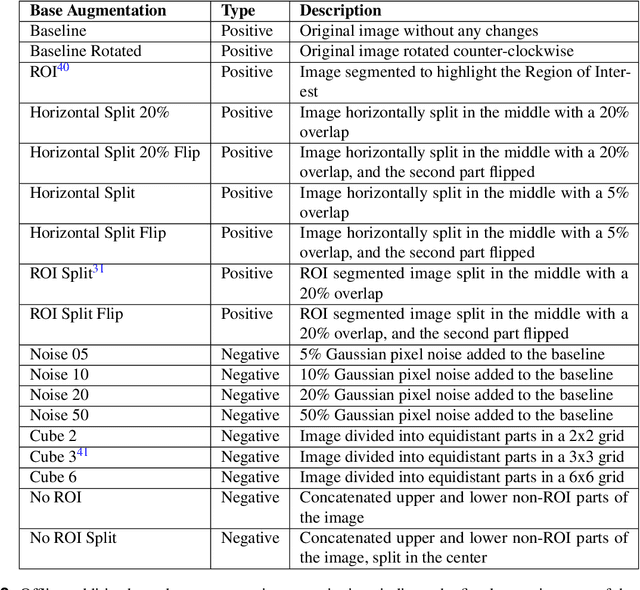
Abstract:Diagnosing knee joint osteoarthritis (KOA), a major cause of disability worldwide, is challenging due to subtle radiographic indicators and the varied progression of the disease. Using deep learning for KOA diagnosis requires broad, comprehensive datasets. However, obtaining these datasets poses significant challenges due to patient privacy concerns and data collection restrictions. Additive data augmentation, which enhances data variability, emerges as a promising solution. Yet, it's unclear which augmentation techniques are most effective for KOA. This study explored various data augmentation methods, including adversarial augmentations, and their impact on KOA classification model performance. While some techniques improved performance, others commonly used underperformed. We identified potential confounding regions within the images using adversarial augmentation. This was evidenced by our models' ability to classify KL0 and KL4 grades accurately, with the knee joint omitted. This observation suggested a model bias, which might leverage unrelated features for classification currently present in radiographs. Interestingly, removing the knee joint also led to an unexpected improvement in KL1 classification accuracy. To better visualize these paradoxical effects, we employed Grad-CAM, highlighting the associated regions. Our study underscores the need for careful technique selection for improved model performance and identifying and managing potential confounding regions in radiographic KOA deep learning.
Improving Performance in Colorectal Cancer Histology Decomposition using Deep and Ensemble Machine Learning
Oct 25, 2023



Abstract:In routine colorectal cancer management, histologic samples stained with hematoxylin and eosin are commonly used. Nonetheless, their potential for defining objective biomarkers for patient stratification and treatment selection is still being explored. The current gold standard relies on expensive and time-consuming genetic tests. However, recent research highlights the potential of convolutional neural networks (CNNs) in facilitating the extraction of clinically relevant biomarkers from these readily available images. These CNN-based biomarkers can predict patient outcomes comparably to golden standards, with the added advantages of speed, automation, and minimal cost. The predictive potential of CNN-based biomarkers fundamentally relies on the ability of convolutional neural networks (CNNs) to classify diverse tissue types from whole slide microscope images accurately. Consequently, enhancing the accuracy of tissue class decomposition is critical to amplifying the prognostic potential of imaging-based biomarkers. This study introduces a hybrid Deep and ensemble machine learning model that surpassed all preceding solutions for this classification task. Our model achieved 96.74% accuracy on the external test set and 99.89% on the internal test set. Recognizing the potential of these models in advancing the task, we have made them publicly available for further research and development.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge