Lawrence J. Bonassar
A watershed-based algorithm to segment and classify cells in fluorescence microscopy images
Jun 02, 2017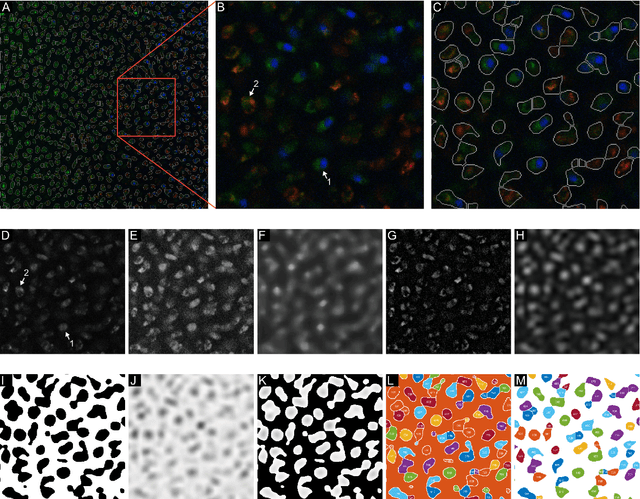
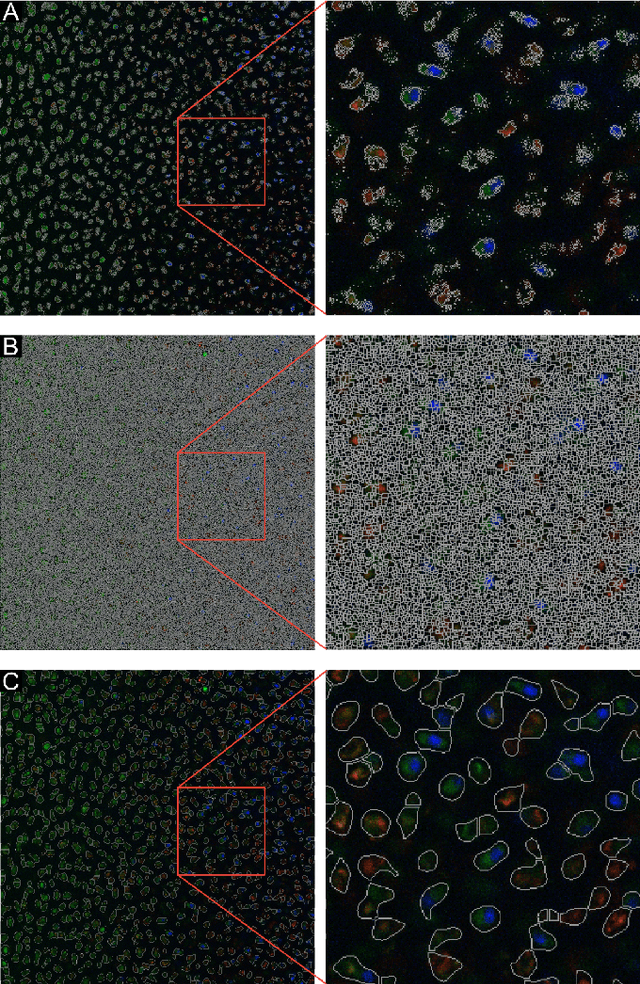
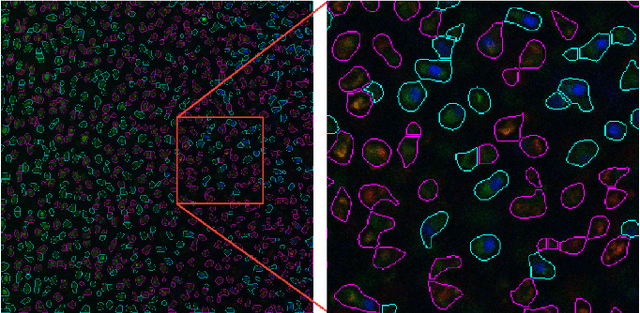
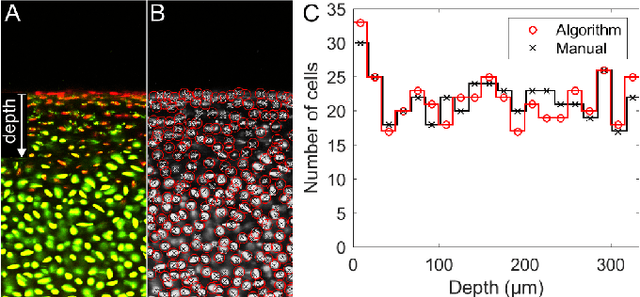
Abstract:Imaging assays of cellular function, especially those using fluorescent stains, are ubiquitous in the biological and medical sciences. Despite advances in computer vision, such images are often analyzed using only manual or rudimentary automated processes. Watershed-based segmentation is an effective technique for identifying objects in images; it outperforms commonly used image analysis methods, but requires familiarity with computer-vision techniques to be applied successfully. In this report, we present and implement a watershed-based image analysis and classification algorithm in a GUI, enabling a broad set of users to easily understand the algorithm and adjust the parameters to their specific needs. As an example, we implement this algorithm to find and classify cells in a complex imaging assay for mitochondrial function. In a second example, we demonstrate a workflow using manual comparisons and receiver operator characteristics to optimize the algorithm parameters for finding live and dead cells in a standard viability assay. Overall, this watershed-based algorithm is more advanced than traditional thresholding and can produce optimized, automated results. By incorporating associated pre-processing steps in the GUI, the algorithm is also easily adjusted, rendering it user-friendly.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge