Konrad U. Förstner
Patent-publication pairs for the detection of knowledge transfer from research to industry: reducing ambiguities with word embeddings and references
Dec 01, 2024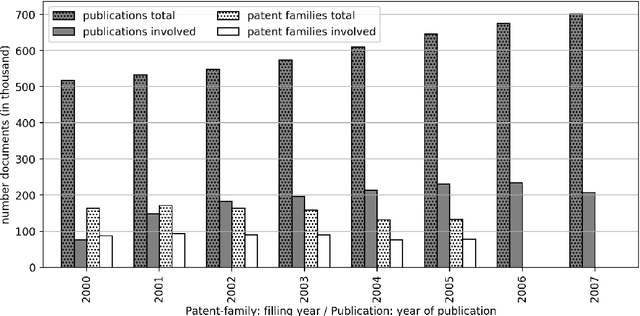
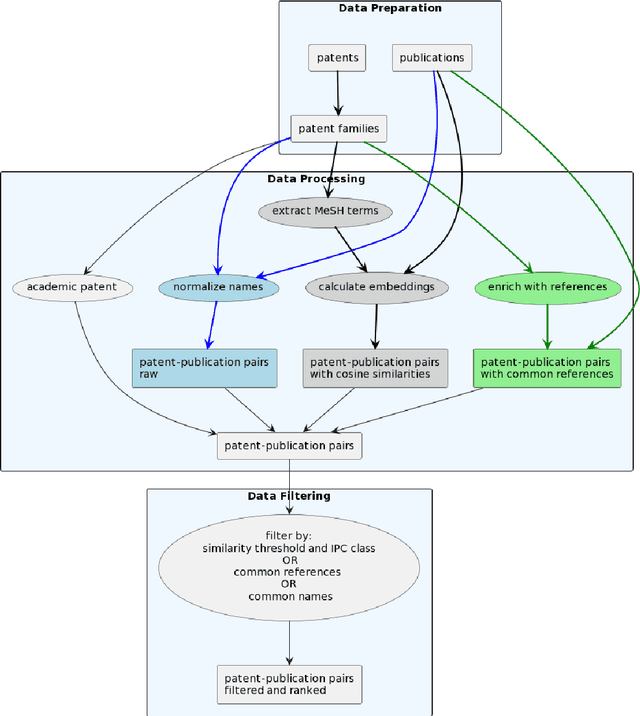
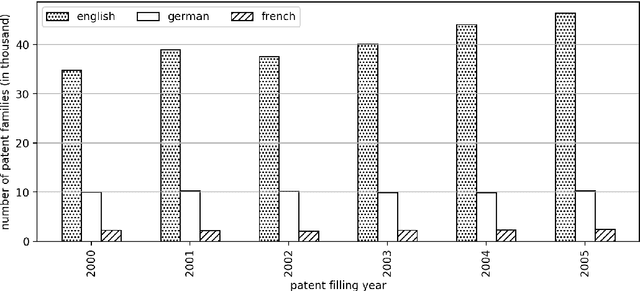
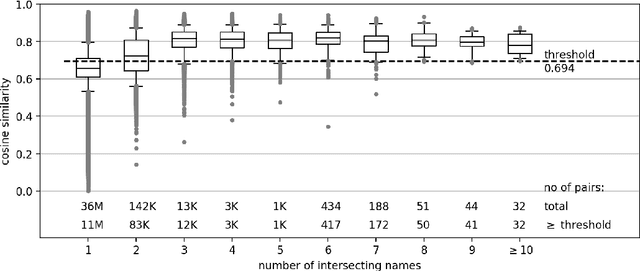
Abstract:The performance of medical research can be viewed and evaluated not only from the perspective of publication output, but also from the perspective of economic exploitability. Patents can represent the exploitation of research results and thus the transfer of knowledge from research to industry. In this study, we set out to identify publication-patent pairs in order to use patents as a proxy for the economic impact of research. To identify these pairs, we matched scholarly publications and patents by comparing the names of authors and investors. To resolve the ambiguities that arise in this name-matching process, we expanded our approach with two additional filter features, one used to assess the similarity of text content, the other to identify common references in the two document types. To evaluate text similarity, we extracted and transformed technical terms from a medical ontology (MeSH) into numerical vectors using word embeddings. We then calculated the results of the two supporting features over an example five-year period. Furthermore, we developed a statistical procedure which can be used to determine valid patent classes for the domain of medicine. Our complete data processing pipeline is freely available, from the raw data of the two document types right through to the validated publication-patent pairs.
Enriched BERT Embeddings for Scholarly Publication Classification
May 07, 2024Abstract:With the rapid expansion of academic literature and the proliferation of preprints, researchers face growing challenges in manually organizing and labeling large volumes of articles. The NSLP 2024 FoRC Shared Task I addresses this challenge organized as a competition. The goal is to develop a classifier capable of predicting one of 123 predefined classes from the Open Research Knowledge Graph (ORKG) taxonomy of research fields for a given article.This paper presents our results. Initially, we enrich the dataset (containing English scholarly articles sourced from ORKG and arXiv), then leverage different pre-trained language Models (PLMs), specifically BERT, and explore their efficacy in transfer learning for this downstream task. Our experiments encompass feature-based and fine-tuned transfer learning approaches using diverse PLMs, optimized for scientific tasks, including SciBERT, SciNCL, and SPECTER2. We conduct hyperparameter tuning and investigate the impact of data augmentation from bibliographic databases such as OpenAlex, Semantic Scholar, and Crossref. Our results demonstrate that fine-tuning pre-trained models substantially enhances classification performance, with SPECTER2 emerging as the most accurate model. Moreover, enriching the dataset with additional metadata improves classification outcomes significantly, especially when integrating information from S2AG, OpenAlex and Crossref. Our best-performing approach achieves a weighted F1-score of 0.7415. Overall, our study contributes to the advancement of reliable automated systems for scholarly publication categorization, offering a potential solution to the laborious manual curation process, thereby facilitating researchers in efficiently locating relevant resources.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge