Khaled Khairy
Z-upscaling: Optical Flow Guided Frame Interpolation for Isotropic Reconstruction of 3D EM Volumes
Oct 09, 2024



Abstract:We propose a novel optical flow based approach to enhance the axial resolution of anisotropic 3D EM volumes to achieve isotropic 3D reconstruction. Assuming spatial continuity of 3D biological structures in well aligned EM volumes, we reasoned that optical flow estimation techniques, often applied for temporal resolution enhancement in videos, can be utilized. Pixel level motion is estimated between neighboring 2D slices along z, using spatial gradient flow estimates to interpolate and generate new 2D slices resulting in isotropic voxels. We leverage recent state-of-the-art learning methods for video frame interpolation and transfer learning techniques, and demonstrate the success of our approach on publicly available ultrastructure EM volumes.
Deep-learning-based acceleration of MRI for radiotherapy planning of pediatric patients with brain tumors
Nov 22, 2023Abstract:Magnetic Resonance Imaging (MRI) is a non-invasive diagnostic and radiotherapy (RT) planning tool, offering detailed insights into the anatomy of the human body. The extensive scan time is stressful for patients, who must remain motionless in a prolonged imaging procedure that prioritizes reduction of imaging artifacts. This is challenging for pediatric patients who may require measures for managing voluntary motions such as anesthesia. Several computational approaches reduce scan time (fast MRI), by recording fewer measurements and digitally recovering full information via post-acquisition reconstruction. However, most fast MRI approaches were developed for diagnostic imaging, without addressing reconstruction challenges specific to RT planning. In this work, we developed a deep learning-based method (DeepMRIRec) for MRI reconstruction from undersampled data acquired with RT-specific receiver coil arrangements. We evaluated our method against fully sampled data of T1-weighted MR images acquired from 73 children with brain tumors/surgical beds using loop and posterior coils (12 channels), with and without applying virtual compression of coil elements. DeepMRIRec reduced scanning time by a factor of four producing a structural similarity score surpassing the evaluated state-of-the-art method (0.960 vs 0.896), thereby demonstrating its potential for accelerating MRI scanning for RT planning.
Joint Deformable Registration of Large EM Image Volumes: A Matrix Solver Approach
Apr 26, 2018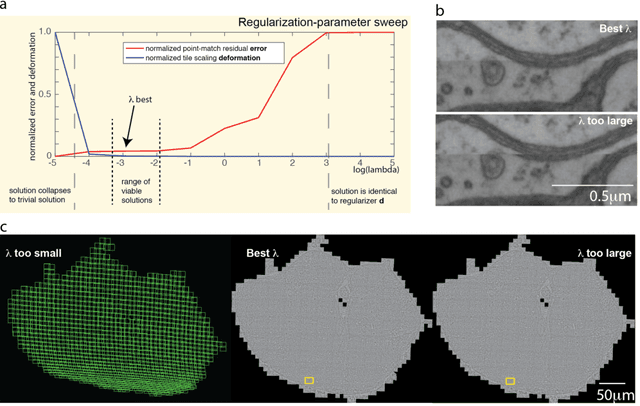

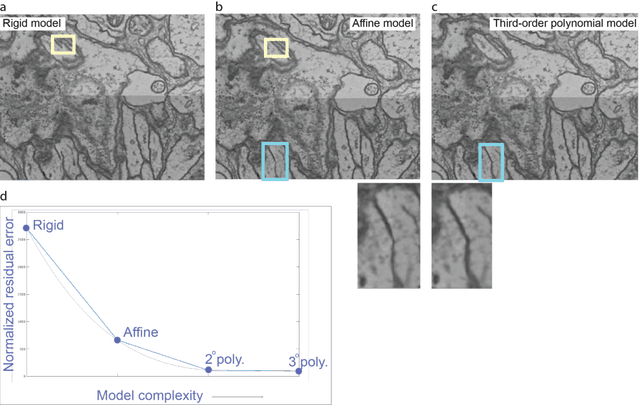
Abstract:Large electron microscopy image datasets for connectomics are typically composed of thousands to millions of partially overlapping two-dimensional images (tiles), which must be registered into a coherent volume prior to further analysis. A common registration strategy is to find matching features between neighboring and overlapping image pairs, followed by a numerical estimation of optimal image deformation using a so-called solver program. Existing solvers are inadequate for large data volumes, and inefficient for small-scale image registration. In this work, an efficient and accurate matrix-based solver method is presented. A linear system is constructed that combines minimization of feature-pair square distances with explicit constraints in a regularization term. In absence of reliable priors for regularization, we show how to construct a rigid-model approximation to use as prior. The linear system is solved using available computer programs, whose performance on typical registration tasks we briefly compare, and to which future scale-up is delegated. Our method is applied to the joint alignment of 2.67 million images, with more than 200 million point-pairs and has been used for successfully aligning the first full adult fruit fly brain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge