Junghwan Cho
Interpretable pap smear cell representation for cervical cancer screening
Nov 17, 2023Abstract:Screening is critical for prevention and early detection of cervical cancer but it is time-consuming and laborious. Supervised deep convolutional neural networks have been developed to automate pap smear screening and the results are promising. However, the interest in using only normal samples to train deep neural networks has increased owing to class imbalance problems and high-labeling costs that are both prevalent in healthcare. In this study, we introduce a method to learn explainable deep cervical cell representations for pap smear cytology images based on one class classification using variational autoencoders. Findings demonstrate that a score can be calculated for cell abnormality without training models with abnormal samples and localize abnormality to interpret our results with a novel metric based on absolute difference in cross entropy in agglomerative clustering. The best model that discriminates squamous cell carcinoma (SCC) from normals gives 0.908 +- 0.003 area under operating characteristic curve (AUC) and one that discriminates high-grade epithelial lesion (HSIL) 0.920 +- 0.002 AUC. Compared to other clustering methods, our method enhances the V-measure and yields higher homogeneity scores, which more effectively isolate different abnormality regions, aiding in the interpretation of our results. Evaluation using in-house and additional open dataset show that our model can discriminate abnormality without the need of additional training of deep models.
Lesion Conditional Image Generation for Improved Segmentation of Intracranial Hemorrhage from CT Images
Mar 30, 2020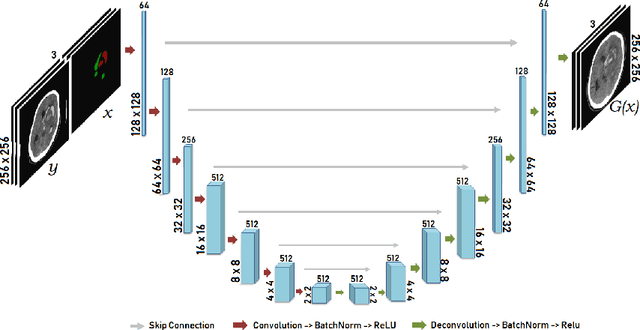
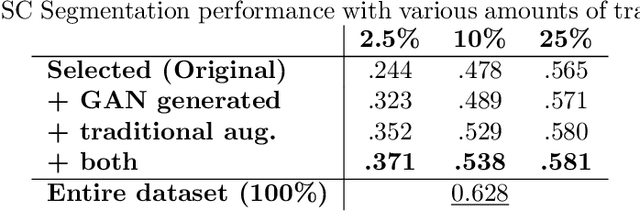

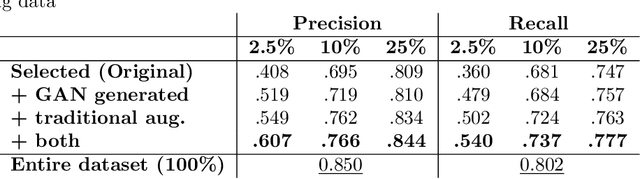
Abstract:Data augmentation can effectively resolve a scarcity of images when training machine-learning algorithms. It can make them more robust to unseen images. We present a lesion conditional Generative Adversarial Network LcGAN to generate synthetic Computed Tomography (CT) images for data augmentation. A lesion conditional image (segmented mask) is an input to both the generator and the discriminator of the LcGAN during training. The trained model generates contextual CT images based on input masks. We quantify the quality of the images by using a fully convolutional network (FCN) score and blurriness. We also train another classification network to select better synthetic images. These synthetic CT images are then augmented to our hemorrhagic lesion segmentation network. By applying this augmentation method on 2.5%, 10% and 25% of original data, segmentation improved by 12.8%, 6% and 1.6% respectively.
How much data is needed to train a medical image deep learning system to achieve necessary high accuracy?
Jan 07, 2016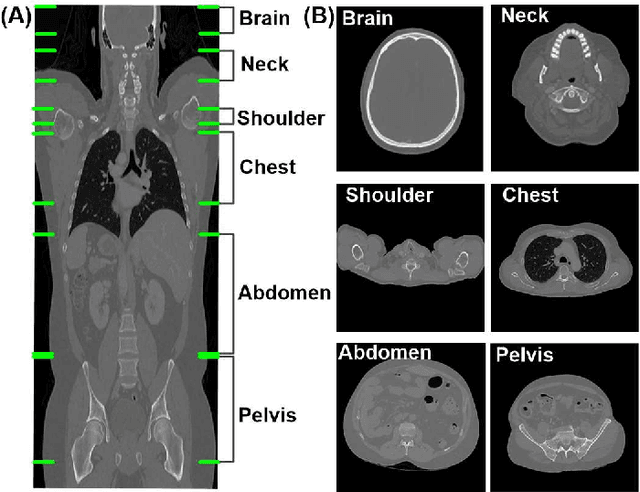
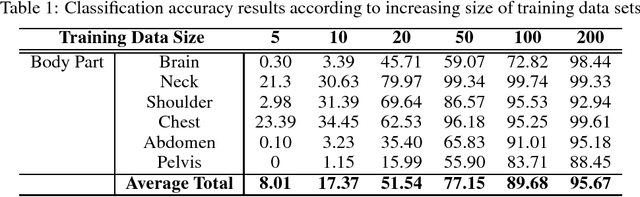
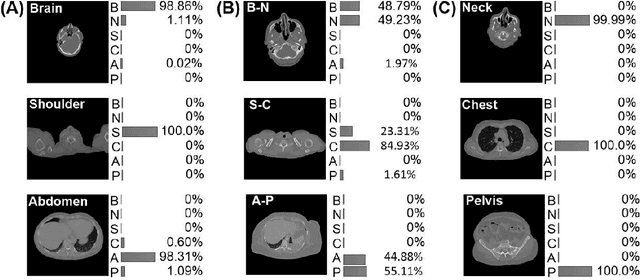
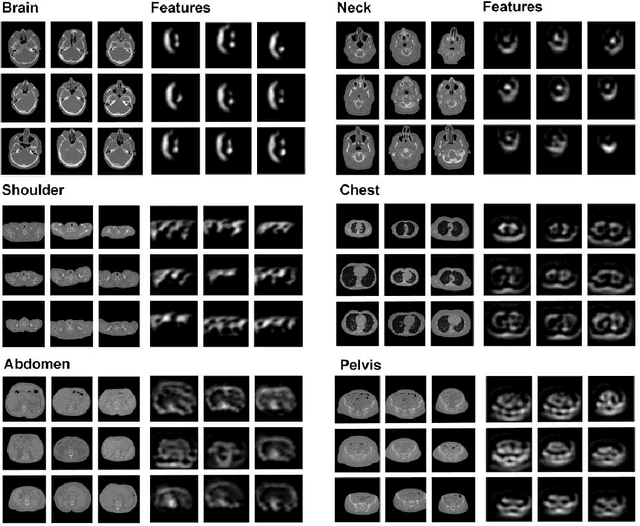
Abstract:The use of Convolutional Neural Networks (CNN) in natural image classification systems has produced very impressive results. Combined with the inherent nature of medical images that make them ideal for deep-learning, further application of such systems to medical image classification holds much promise. However, the usefulness and potential impact of such a system can be completely negated if it does not reach a target accuracy. In this paper, we present a study on determining the optimum size of the training data set necessary to achieve high classification accuracy with low variance in medical image classification systems. The CNN was applied to classify axial Computed Tomography (CT) images into six anatomical classes. We trained the CNN using six different sizes of training data set (5, 10, 20, 50, 100, and 200) and then tested the resulting system with a total of 6000 CT images. All images were acquired from the Massachusetts General Hospital (MGH) Picture Archiving and Communication System (PACS). Using this data, we employ the learning curve approach to predict classification accuracy at a given training sample size. Our research will present a general methodology for determining the training data set size necessary to achieve a certain target classification accuracy that can be easily applied to other problems within such systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge