Jung-Yi Joyce Lin
Variable selection with missing data in both covariates and outcomes: Imputation and machine learning
Apr 06, 2021
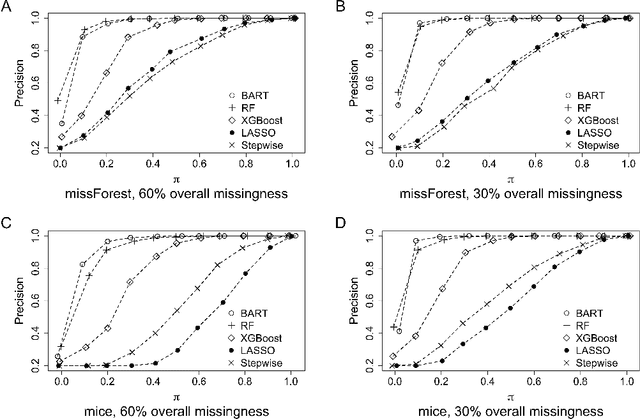


Abstract:The missing data issue is ubiquitous in health studies. Variable selection in the presence of both missing covariates and outcomes is an important statistical research topic but has been less studied. Existing literature focuses on parametric regression techniques that provide direct parameter estimates of the regression model. In practice, parametric regression models are often sub-optimal for variable selection because they are susceptible to misspecification. Machine learning methods considerably weaken the parametric assumptions and increase modeling flexibility, but do not provide as naturally defined variable importance measure as the covariate effect native to parametric models. We investigate a general variable selection approach when both the covariates and outcomes can be missing at random and have general missing data patterns. This approach exploits the flexibility of machine learning modeling techniques and bootstrap imputation, which is amenable to nonparametric methods in which the covariate effects are not directly available. We conduct expansive simulations investigating the practical operating characteristics of the proposed variable selection approach, when combined with four tree-based machine learning methods, XGBoost, Random Forests, Bayesian Additive Regression Trees (BART) and Conditional Random Forests, and two commonly used parametric methods, lasso and backward stepwise selection. Numeric results show XGBoost and BART have the overall best performance across various settings. Guidance for choosing methods appropriate to the structure of the analysis data at hand are discussed. We further demonstrate the methods via a case study of risk factors for 3-year incidence of metabolic syndrome with data from the Study of Women's Health Across the Nation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge